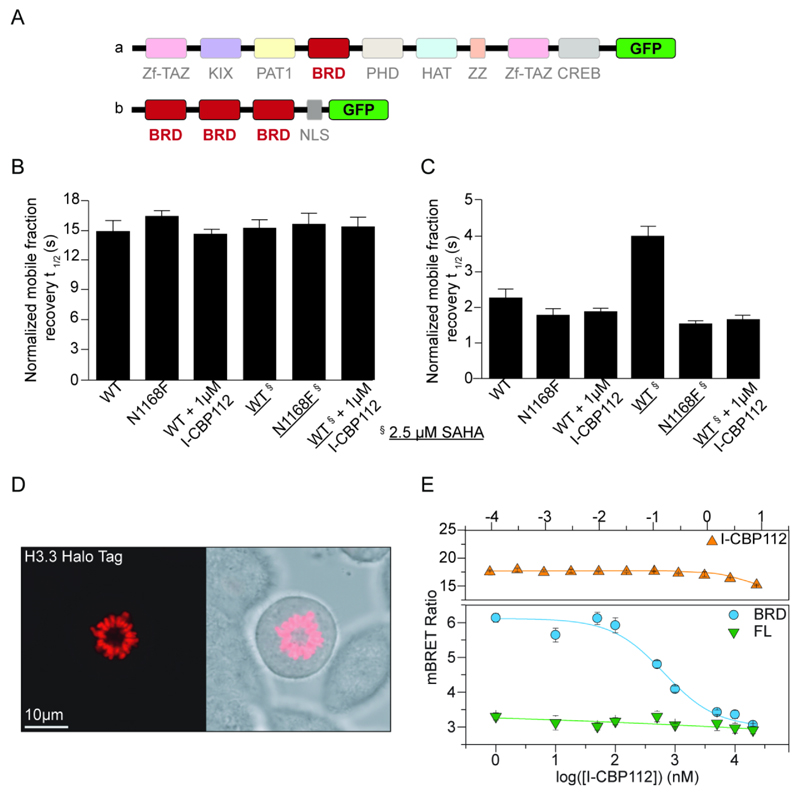
Figure 3. I-CBP112 displaces the CBP bromodomain from chromatin.
A: Domain organization of full length CBP (upper panel) and of the 3xCBPBRD/GFP construct used (lower panel). B: FRAP experiment showing half-recovery times for full length CBP/GFP and its inactivating mutant N1168F. C: FRAP experiment showing half-recovery times of the fluorescent signal of 3xCBPBRD/GFP (WT) and its inactivating mutant . Experiments with SAHA treated cells (2.5 µM) are indicated by underlined constructs. D: Fluorescent image (left panel) and phase contrast image (right panel) of metaphase chromosomes of Halo-tagged histone H3.3 transfected HEK293T cells showing incorporation of the tagged histone into chromatin. E: NanoBRET experiments showing the dose response of NanoLuc Luciferase fusion full-length CBP and the isolated CBP bromodomain. The construct containing only the bromodomain was readily displaced by I-CBP112 with a cellular IC50 value of 600 ± 50 nM. No displacement was observed using full-length GFP-BRD4 in agreement with the in vitro selectivity of I-CBP112 (upper panel).