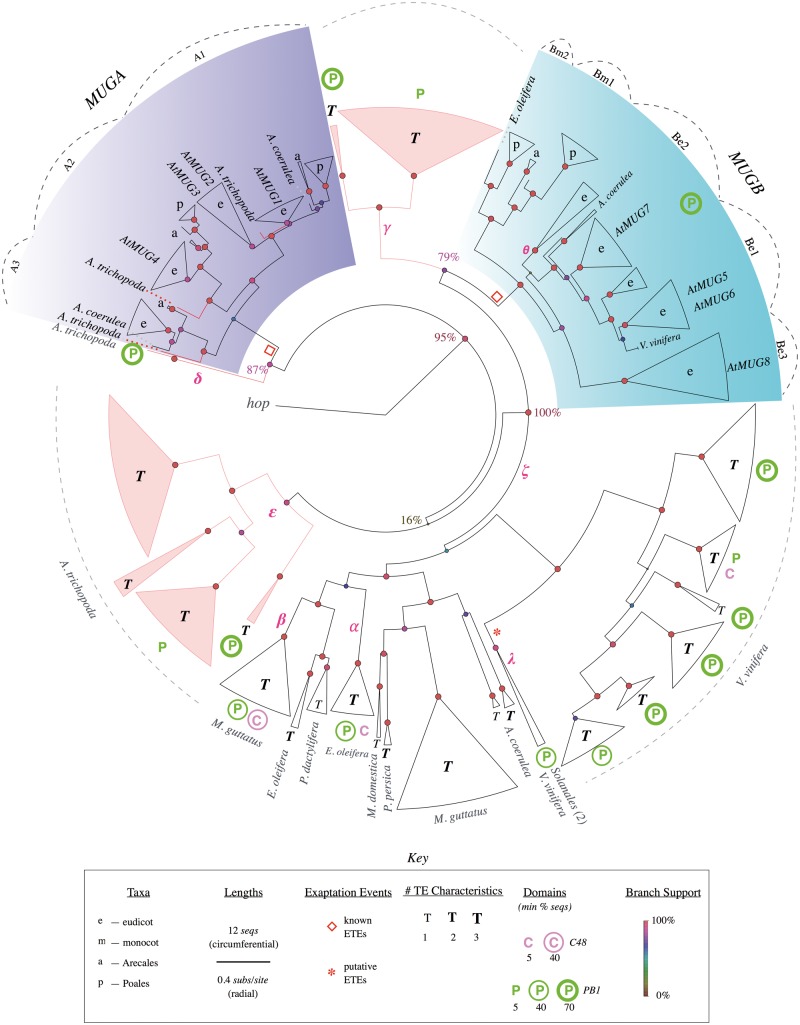
Fig. 2.
Simplified phylogenetic tree of MUG genes and TEs. Curated phylogenetic tree showing sequences descended from the last common ancestor of MUGA and MUGB, rooted at fungal hop. Terminal triangles represent clades, with circumferential width proportional to number of genes (see key). In clades with genes from only one or a few species the species are labeled, otherwise clades are labeled according to taxon. Clades and branches from Amborella trichopoda are colored red. Major clades of MUGA and MUGB are labeled according to Joly-Lopez et al. (2012) and the positions of Arabidopsis thaliana AtMUG1-8 genes are indicated. For simplicity, TE features are categorized by the number of TE characteristics (out of 3) associated with each clade to emphasize differences between clades of known ETEs and clades of putative TEs (see key). For the same tree including detailed TE characteristics, see supplementary figure S3, Supplementary Material online. Radial branch lengths are proportional to the inferred number of substitutions per site (circumferential branch length is arbitrary). Circles at internal nodes have color and size corresponding to “local support values” (Shimodaira–Hasegawa test [Zeh et al. 2009; Oliver et al. 2013]). Empty red diamonds indicate known exaptation events; red asterisks indicate putative novel exaptation events. Greek letters indicate branches referred to in the main text. Dashed lines indicate clades, dotted lines are species labels. Pink branches are used to highlight Am. trichopoda clades or individual sequences. See supplementary figure S2, Supplementary Material online, for a fully expanded phylogenetic tree.