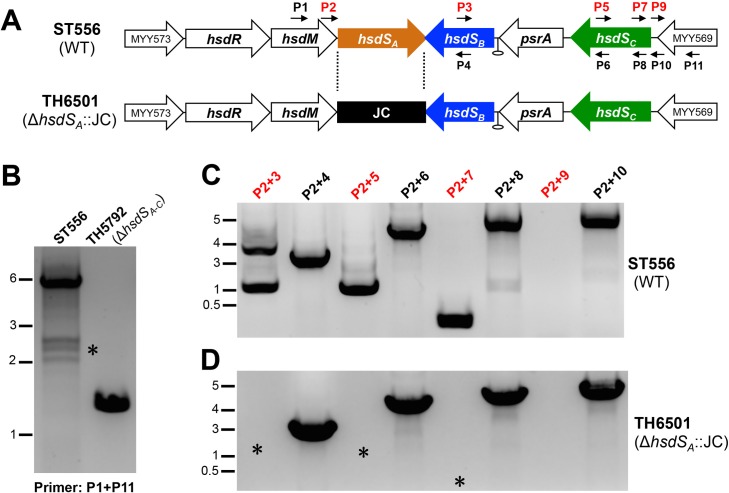
Fig 2. Detection of DNA rearrangements in the Spn556II locus by PCR.
A. Positions of the primers used for PCR amplification in the Spn556II locus of ST556. The predicted rho-independent transcription terminator is indicated by a hairpin. The primers used in (B) and (C) are indicated by small arrows. The JC-replaced region in TH6501 is marked with dashed lines. B. Amplification of the Spn556II locus in ST556 and isogenic mutant TH5792 lacking the entire Spn556II locus with primers P1 and P11. The PCR mixtures were processed by DNA electrophoresis and stained by the Goldview dye (Yeasen, Beijing, China). The PCR products that were absent in the mutant strains are marked with asterisks (*). The sizes of the DNA markers are indicated in kilobases. C. Detection of DNA rearrangements in the hsdS regions of the Spn556II locus. PCR reactions were performed with the genomic DNA of ST556 using the same set of primer pairs indicated at the top of each lane, and marked as in (B). D. Same as in (C) except for using the genomic DNA from the ST556 derivative lacking hsdS A strain (TH6501).