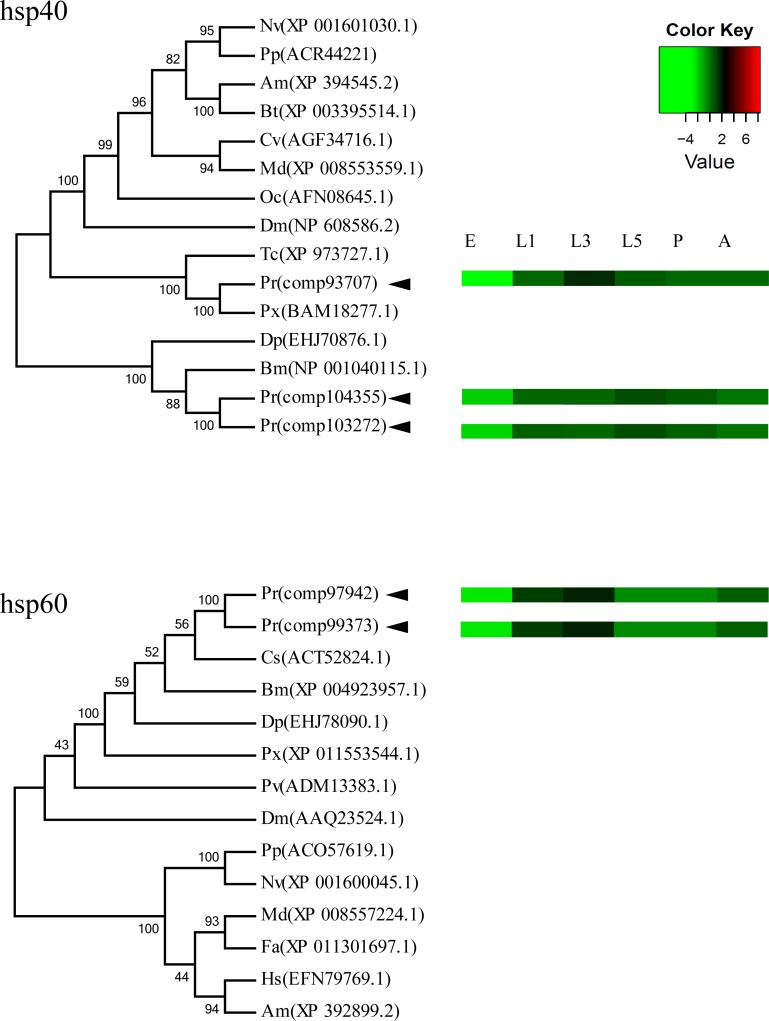
Fig 6. The homology relationships of Pieris rapae heat shcok proteins with their counterparts in other insect species as well as expression levels in P. rapae during development.
The trees were generated using the NJ method in Mega5 software. Expression levels were compared with their RPKM values. The other Hsp amino acid sequences used are from Nv (Nasonia vitripennis), Am (Apis mellifera), Bt (Bombus terrestris), Cv (Cotesia vestalis), Md (Microplitis demolitor), Oc (Oxya chinensis), Dm (Drosophila melanogaster), Tc (Tribolium castaneum), Pr (Pieris rapae), Px (Papilio xuthus), Dp (Danaus plexippus), Bm (Bombyx mori), Cs (Chilo suppressalis), Pv (Polypedilum vanderplanki), Fa (Fopius arisanus), and Hs (Harpegnathos saltator).