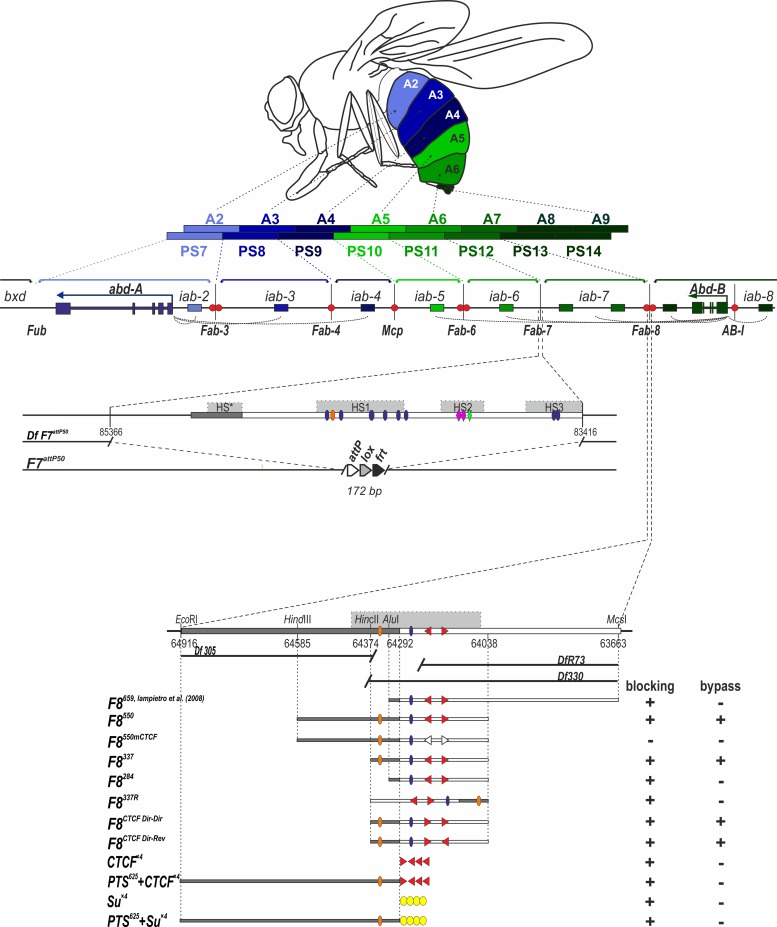
Fig 1. Fragments of Fab-8 used for Fab-7 replacement.
(A) Regulatory region of the distal part of the BX-C. Horizontal arrows represent transcripts for abd-A (blue) and Abd-B (green). iab enhancers are shown as rectangles color-coded to the respective gene they control (darker shades of color indicate higher expression levels). The dotted arches are a graphical illustration of the targeting of each cis-regulatory domain to the abd-A or Abd-Bm promoter. Vertical lines mark boundaries (Fub, Fab-3, Fab-4, Mcp, Fab-6, Fab-7, and Fab-8) of regulatory iab domains which are delimited by brackets above the map. There is also a boundary-like element AB-I upstream of the Abd-B promoter that has communicator activity in bypass assays. CTCF binding sites at boundaries are shown as red circles. Abdominal segments (A2-9) and parasegments (PS7-14) that correspond to the iab domains of abd-A and Abd-B are shown as multicolored bars. Localization of the segments in adult male is shown on the fly drawing at the top. Deletion of Fab-7 in Fab-7attP50 is shown separately. (B) Molecular maps of the Fab-8 boundary replacement constructs analyzed in the paper and their blocking and bypass activities. Fab-8 insulator is shown as a horizontal bar. PTS is marked by dark gray. DNase hypersensitive site is shown as a light gray box above the coordinate bar. The proximal and distal deficiency endpoints of the Fab-8 deletions are shown below. Known protein binding sites are indicated. Binding factors, common for Fab-7 and Fab-8, are shown as ovals: blue–GAF, orange–Elba/Insv. The non-common factors for Fab-7 –as rhombi: rose–Pita, green–Zipic. dCTCF binding sites are shown as red triangles indicating orientation of the sites. Empty triangles mark the mutated sites. Su(Hw) binding sites are denoted as yellow ovals. On the right side of the constructs, the blocking and bypass activities of each replacement construct are shown.