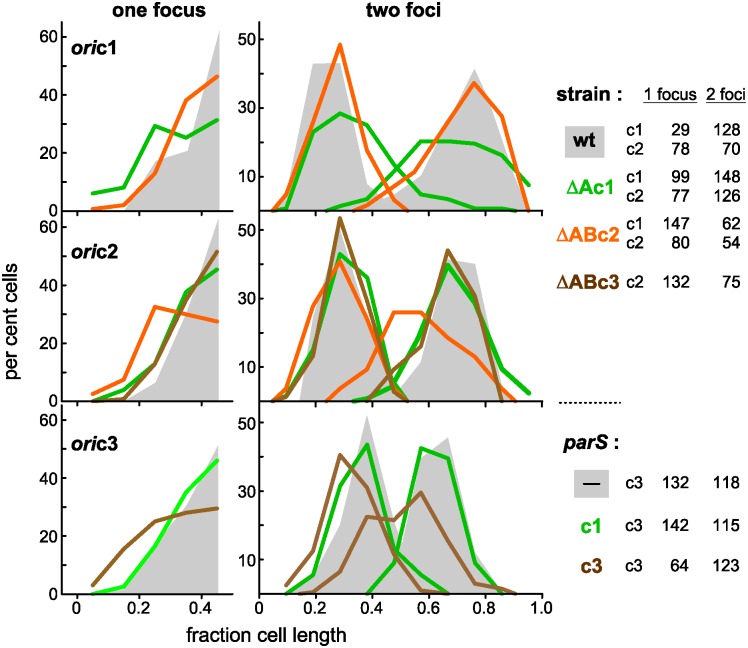
Fig 5. Effect of ParABS disruption on distribution of replication origins.
The chromosome origins of Δpar and corresponding wild type strains were marked with ParB::Fp proteins, and the distances of fluorescent foci from the most focus-proximal pole were measured and binned in intervals of one-tenth cell length. The ParABS disrupting factors are shown at the right: deletion mutations in oric1- and oric2-marked cells (grown in MglyC), plasmid-borne parS sites in oric3-marked cells (grown in SOB). Origin distributions in cells with undisturbed Par function are shown as shaded areas. Numbers of one- and two-focus cells analyzed are shown at the right.