Abstract
Monocytes and macrophages are important components of the immune system, specialized in either removing pathogens as part of innate immunity or contributing to adaptive immunity through antigen presentation. Essential to such functions is classical activation (M1) and alternative activation (M2) of macrophages. M1 polarization of macrophages is characterized by production of pro-inflammatory cytokines, antimicrobial and tumoricidal activity, whereas M2 polarization of macrophages is linked to immunosuppression, tumorigenesis, wound repair and elimination of parasites. MiRNAs are small non-coding RNAs with the ability to regulate gene expression and network of cellular processes. A number of studies have determined miRNA expression profiles in M1 and M2 polarized human and murine macrophages using microarray and RT-qPCR arrays techniques. More specifically, miR-9, miR-127, miR-155 and miR-125b have been shown to promote M1 polarization while miR-124, miR-223, miR-34a, let-7c, miR-132, miR-146a and miR-125a-5p induce M2 polarization in macrophages by targeting various transcription factors and adaptor proteins. Further, M1 and M2 phenotypes play distinctive roles in cell growth and progression of inflammation-related diseases such as sepsis, obesity, cancer and multiple sclerosis. Hence, miRNAs that modulate macrophage polarization may have therapeutic potential in the treatment of inflammation-related diseases. This review highlights recent findings in miRNA expression profiles in polarized macrophages from murine and human sources, and summarizes how these miRNAs regulate macrophage polarization. Lastly, therapeutic potential of miRNAs in inflammation-related diseases through modulation of macrophage polarization is also discussed.
Keywords: microRNAs, inflammation, monocytes, macrophage polarization, sepsis
1. Introduction
Monocytes are derived from common myeloid progenitor cells in bone marrow. From there, they enter the peripheral blood stream and continually migrate into all tissues, where they differentiate into macrophages, a mature form of monocytes (1). Macrophages are relatively long-lived cells with many important functions. One is to eliminate invading microorganisms via phagocytosis, which is a critical first defense in innate immunity (2). Another crucial role of macrophages is to modulate adaptive immune response to various pathogens through antigen processing and presentation (2). They would either induce inflammation or repair damaged tissues by secreting different signaling proteins. In addition to these functions in the immune system, macrophages can act as scavenger cells for clearing dead cells, cell debris, and other molecules such as cholesterol and fatty acids (2). Different functions of macrophages are associated with the type of receptor interaction and the presence of cytokines (2). In recent years, the concept of classical (M1) and alternative (M2) activation of macrophages has become increasingly recognized. The classical M1 macrophages are activated by “pro-inflammatory” cytokine profiles (i.e. IL-6 and TNF-α), but the alternative M2 macrophages appear to be involved in immunosuppression and tissue repair with low or null levels of pro-inflammatory cytokine secretion (3).
MicroRNAs (miRNAs) are small, endogenous, non-coding RNAs of ∼22 to 26 nucleotides in length that function primarily as post-transcriptional regulators. Accumulating evidences have shown that miRNAs can regulate many pathophysiological processes such as cell proliferation, metabolism, apoptosis, and organ development (4). Accordingly, miRNAs have been shown to regulate macrophage polarization and subsequent effects on inflammation (5). In this review, we will summarize recent findings in miRNA profiles in human and mouse polarized macrophages, and discuss the mechanisms underlying the miRNA-mediated regulation of macrophage polarization. We will also discuss the therapeutic potential of miRNA-mediated macrophage polarization in inflammation-related disease (i.e., sepsis, obesity, cancer, multiple sclerosis).
2. MiRNA biogenesis and mechanism of action
The conventional miRNA biogenesis is initiated by the transcription of genes (mostly from the intron regions) by the type II RNA polymerase in the nucleus, generating primary miRNA (pri-miRNA), which will be cleaved by the endoribonuclease Drosha/DGCR8 complex and yield precursor miRNA (pre-miRNA) (6). Exportin 5 is the major transporter that exports pre-miRNA to the cytoplasm, where mature miRNA is formed via cleaving the loop end of the pre-miRNA by the Dicer complex. Together with other molecules such as Argonaute 2, mature miRNA is assembled into the miRNA-induced silencing complex, which would recognize and bind to the 3′ untranslated region (3′ UTR) of target mRNAs using the seed sequence through Watson-Crick base-pairing mechanism (6). MiRNA suppresses gene expression by causing target mRNA instability/degradation when perfect base-pairing occurs, or by inhibiting translation when it forms imperfect complementary base pairs (6). Recent evidence has indicated that the latter one is the primary mechanism by which miRNAs affect target gene expression (7). Interestingly, mature miRNA was previously denoted as the guide strand with inhibitory function, whereas another strand (miRNA*) was regarded as the passenger strand that had no cellular activity and would be degraded after formation. However, recent findings have demonstrated that the passenger strand is functional in regulating cellular process like the guide strand. For instance, miR-223 (guide strand, -3p) and miR-223* (passenger strand, -5p) were both dysregulated in mouse hearts with moderate or severe sepsis and loss of miR-223 duplex (3p and 5p) exacerbated myocardial dysfunction and mortality caused by sepsis (8).
In addition to the activity within the cells in which they are transcribed, miRNAs can be secreted into extracellular compartments and delivered to distant target tissues. These miRNAs, along with other contents such as proteins or other RNAs (mRNAs, circRNAs, and lncRNAs), can be encapsulated into exosomes or microvesicles (4). Upon conventional uptake of exosomal content via direct fusion of cell membrane or endocytosis, miRNAs can regulate gene expression at post-transcriptional level and affect normal cellular activity in the recipient cell (4, 9). This novel mechanism of miRNA transfer constitutes an important element of intracellular communication. Very recently, our lab has reported that stem cell-derived exosomal miR-223 significantly suppressed the production of pro-inflammatory cytokines (TNF-α, IL-1β, and IL-6) by targeting Sema3A and STAT3 in macrophages of septic model (10). Similarly, another study showed that miR-155 and miR-146a, present in dendritic cell-derived exosomes, were passed between immune cells in vivo (11). Exosomal miR-146a inhibited while miR-155 promoted endotoxin-induced inflammation in mice (11). Currently, it is unclear whether exosomal miRNAs enter the nucleus of the target cell.
3. Mechanisms of macrophage polarization
3.1 M1 activation
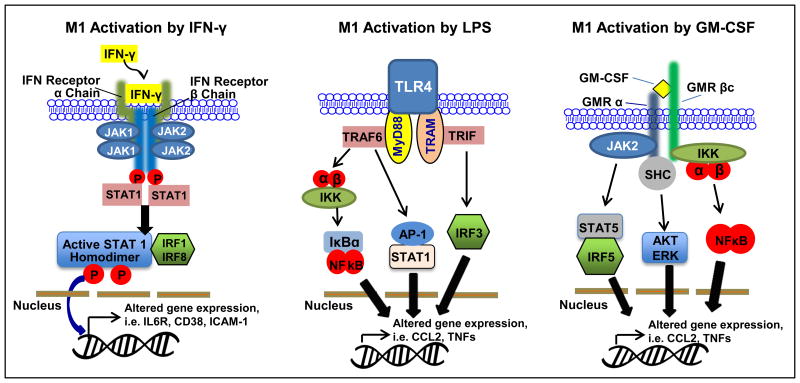
Classical activation (M1) transforms macrophages into potent antimicrobial effector cells, and it requires two signals that can be fulfilled by the effector Th1 cells. One signal is the cytokine IFN-γ while the other signal, CD40 ligands, sensitizes macrophages to respond to IFN-γ (12, 13). Other cells, such as natural killer (NK) cells or macrophages themselves, have been shown to produce such cytokine as well. Once IFN-γ binds to the receptor (IFNGR), it recruits Janus kinase 1 (Jak1) and Jak2 adaptors, which activate factors (IRF1 or IRF8). This signaling pathway regulates specific gene expression of cytokine receptors (IL6R, IL2RA, or IL15 receptor α), cell activation markers (CD38, CD69 or CD97) and cell adhesion molecules (ICAM1, integrin α L, and mucin 1). Upon stimulation by these molecules, macrophage secretes TNF-α, which further stimulates macrophages through the TNFR1. Classical activation of macrophages would then increase the destruction of engulfed pathogens (14-16) (Fig. 1).
Figure 1. Classical activation of macrophages (M1).
Ligands IFN-γ, LPS and GM-CSF activate STAT1, NF-κB, AP-1, IRFs to stimulate transcription of M1 associated genes.
Other markers and M1 stimuli that induce prototypic inflammatory responses may originate from different sources and activate distinct signaling pathways. Two major examples are the bacterial lipopolysaccharide (LPS) and granulocyte macrophage colony stimulating factor (GM-CSF) (17). LPS is the well-known M1 macrophage signal that is propagated by the toll-like receptor 4 (TLR4) cascade (17). Similar to IFN-γ signaling pathway, LPS-mediated M1 activation is under the control of NF-κB, activator protein 1 (AP-1), IRFs, and STAT1. Typically, TLR4 activation leads to strong pro-inflammatory cytokine signaling (i.e. IL-1β, IL-6, IL-12, TNF, and IFN-β), high levels of chemokine secretion [i.e. Chemokine (C-C motif) ligand 2 (CCL2), chemokine (C-X-C motif) ligand 10 (CXCL10), and CXCL11], and antigen presenting molecules such as MHC family members (17) (Fig. 1).
GM-CSF is defined as a hematopoietic growth factor that originates from various cell types including T cells, macrophages, endothelial cells and fibroblasts upon different stimuli (18). The pro-inflammatory signal pathway of GM-CSF is very similar to IFN-γ and LPS. Activation of GM-CSF receptor (GMR) recruits JAK2, which activates STAT5, extracellular signal-regulated kinase (ERK), NF-κB and IRF5 (18). For the most part, secretion of pro-inflammatory cytokines (i.e. IL-1β, Il-6, TNF-α) are lower in GM-CSF-stimulated macrophages compared to LPS-induced macrophages, although fold expressions are not always consistent for the listed cytokines (18) (Fig. 1).
3.2 M2 activation
In contrast, alternative macrophage polarization is characterized by its ability to antagonize prototypic inflammatory responses by helping parasite clearance and promoting tissue remodeling (19). However, the mechanism of activation is more complicated and does not constitute a uniform population. Based on different stimuli, M2 macrophages can be further categorized into subsets: M2a, M2b, M2c, and M2d (19). Each category displays unique gene expression profile, but production of high levels of IL-10 and IL-1 receptor antagonist (IL-1RA) and low levels of IL-12 are observed in all subsets (19, 20). One key signature of all M2 subsets is the production of Arginase-1 enzyme that depletes L-arginine, which is the substrate of inducible nitric oxide synthase (iNOS), leading to suppression of T cell responses (19).
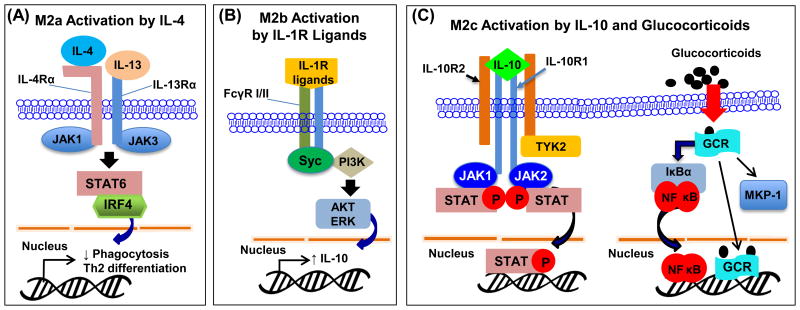
M2a is elicited by IL-4 (or in some cases, IL-13) that is produced by Th2 cells, eosinophils, basophils, and macrophages (21). Upon binding to receptors (IL-4Rα1, IL-13Rα1, or IL-13Rα2), JAK1 and JAK3 are activated, leading to activation of STAT6 and IRF4. IL-4-induced macrophage polarization would then decrease phagocytosis activity (Fig. 2A). Knocking out IL-4 does not affect the number and maturation of macrophages, but impairs immune response against nematodes and viral infections (21).
Figure 2.
(A) M2a polarization. Ligands IL-4 and IL-13 activate STAT6/IRF4 signaling pathway to induce M2a phenotype in macrophages. (B) M2b polarization. IL-1R ligands or immune complexes in combination with LPS activate Syc and PI3K to induce transcription of IL-10 and M2b phenotype. (C) M2c polarization. IL-10 and glucocorticoids activate M2c phenotype through STAT, NF-κB and MKP-1 activation.
The type II-activated macrophages (M2b) are triggered by IL-1R ligands or exposure to immune complexes plus LPS (21). These ligands are recognized by Fc receptor family on the cell membrane, including activatory FcγR1 (or CD64) and inhibitory FCγRIIA (CD32). The signal is then passed down to the spleen tyrosine kinase (Syc) and phosphoinositide 3-kinase (PI3K), which increases IL-10 and TNF-α expression (21) (Fig. 2B).
Glucocorticoids (GC) and IL-10 are the main mediators that trigger M2c subtype phenotype (13). Glucocorticoids diffuse into the cell and bind to its receptor (GCR), followed by translocation of the complex to the nucleus. This complex can interact with NF-κB and AP-1, thereby interfering with inflammatory responses (22, 23). Moreover, it is reported that GCR impairs p38 MAPK activity by inducing MAP kinase phosphatase-1 (MKP-1), leading to improved survival outcome from LPS-induced mortality (24). The chemokine expression profile differs from other M2 subtypes with high expression of CCR2, which affect monocyte adherence, phagocytosis, and apoptosis (25, 26) (Fig. 2C).
Tumor-associated macrophages (TAMs) are the major inflammatory component of the tumor microenvironment (TME). Since they share the similar IL-10highIL-12low phenotype as M2 macrophages, TAMs are categorized as a novel M2 subset – M2d.(20, 27). TAMs can be activated by IL-6 and macrophage colony stimulating factor (M-CSF). Upon Fra-1 binding to the IL-6 promoter in macrophages, increased production of IL-6 acts in an autocrine manner to induce M2d polarization (27). In addition, leukemia inhibitory factor (LIF) has been identified as another micro-environmental factor that promotes TAM transformation (28). Indeed, LIF and IL-6 can facilitate consumption of M-CSF by monocytes, leading to the differentiation of monocytes to TAM-like cells (28). Depletion of LIF, IL-6 and M-CSF suppresses such induction of TAM-like cells (28). TAMs share similar cytokine profiles with other M2 subtypes with augmented production of IL-10, TGF-β (29). However, the chemokine signatures of TAMs are distinct from other M2 subtypes by secretion of high levels of CCL5, CXCL10 and CXCL16 (29). Interestingly, TAMs also express M1 markers and can be transformed into M1-like phenotype with LPS or IFN-γ stimulation (30, 31).
4. MicroRNAs profiles in polarized macrophages of human and mice
A number of studies have profiled the expression of miRNAs in the various polarizing conditions in human- and murine-derived macrophages. Zhang et al. (32) used murine bone marrow-derived macrophages (BMDMs) to determine miRNA expression in M1 and M2 polarizing conditions. In this study, M1-polarized conditions in BMDMs were induced by LPS plus IFN-γ treatment whereas M2 polarization was stimulated by IL-4 (32). The miRNA-microarray results revealed that expression levels of 109 miRNAs were altered between M1 and M2 conditions (32). Using qRT-PCR, they confirmed that in M1 polarized macrophages, miR-181a, miR-155-5p, miR-204-5p and miR-451 were upregulated, whereas miR-125-5p, miR-146a-3p, miR-143-3p and miR-145-5p were downregulated in comparison to those levels in M2 polarized conditions (32). Although the qRT-PCR results of some miRNAs were inconsistent with the microarray results, the majority of miRNAs analyzed were congruent in both assays.
On the other hand, Cobos Jimenez et al. (33) performed miRNA profile assays in polarized monocytes derived from human peripheral blood mononuclear cells (PBMCs). They observed that 303 miRNAs were upregulated or downregulated in M1 (induced by combination of IFN-γ and TNF-α), M2a (induced by IL-4) or M2c (induced by IL-10) conditions compared to unstimulated conditions after miRNA arrays (33). They further confirmed, using RT-PCR, that miR-125b-5p, miR-181a- 5p, miR-193b-3p and miR-125a-5p were significantly upregulated in M1 macrophages compared to unstimulated conditions but these miRNAs were also dysregulated in M2 conditions (33). MiR-145-5p, miR-146a-5p, miR-193a-5p and miR-29b-3p were upregulated whereas miR-629-5p was downregulated solely in M1 macrophages (33). MiR-500a-5p and miR-502–3p were upregulated while miR-181b-5p was downregulated only in M2a macrophages (33). Also, miR-21- 5p, miR-22-3p and miR146b-5p were upregulated whereas miR-339-3p and miR-200a-3p were downregulated solely in M2c conditions (33).
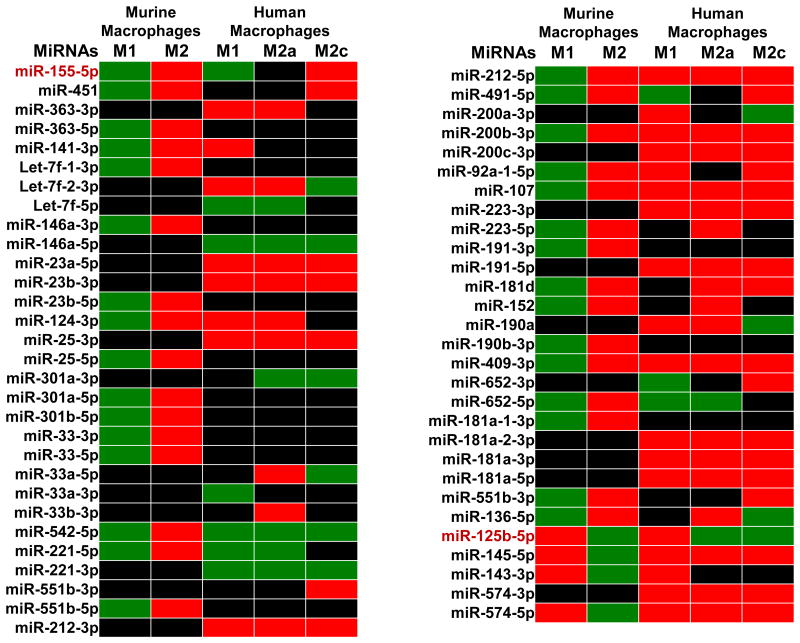
Figure 3 summarizes a comparison of miRNA profiles in polarized macrophages from murine and human sources as presented by Zhang et al. (32) and Cobos Jimenez et al. (33). As showed in Figure 3, only miR-155-5p and miR-125b-5p showed similar expression patterns in murine and human polarized macrophages across both M1 and M2 polarized conditions. Some miRNAs displayed consistent expression profiles in human and murine macrophages in a single polarized condition but had opposite profiles in the other. For instance, miR-221-5p was upregulated in both murine and human M1 macrophages but was downregulated in murine M2 macrophages, which contradicted with upregulation of levels in human M2a macrophages. Differences in origin of macrophages used in these studies may account for the contrasting expression profiles. Generally, human macrophages used in studies are isolated from blood peripheral mononuclear cells whereas tissue-associated or bone marrow-derived macrophages are utilized in murine studies (34). Thus, miRNA expression profiles may differ in polarized conditions depending on the location and type of macrophage. This idea expounds on recent reports that adult tissue-resident macrophages do not originate from bone marrow (35).
Figure 3.
miRNA expression profile in polarized macrophages, based on the papers by Zhang et al. (28) and by Cobos Jimenez et al. (29). Green: Upregulated miRNAs, Red: Downregulated miRNAs, Black: unknown or unchanged miRNAs.
In the same light, Graff et al. (36) compared expression of miRNAs in polarized human monocyte-derived macrophages (MDM) versus polarized human monocytic THP-1 cell line. They observed that miR-125a-3p was upregulated in M1 polarized conditions, miR-193b in M2a conditions and miR-27a* in M2b conditions. Also, miR-155* was increased in both M1 and M2b conditions. Consistently, miR-125a-3p, miR-27a* and miR-155* had similar expression profiles in polarized THP-1 monocytes as in polarized MDMs. Luciferase assays also revealed that miR-29b and miR125a-5p targeted TNFAIP3, a negative regulator of NF-κB signaling (36).
In summary, profiling miRNA expression in polarized macrophages with techniques such as microarray and RT-qPCR arrays yields large amounts of dysregulated miRNAs. For the most part, the functional roles of such dysregulated miRNAs, in the studies discussed above, in regulating macrophage polarization were unexplored. Nonetheless, some studies, discussed below, have investigated the precise roles of miRNAs in the activation and polarization of macrophages.
5. Functional role of microRNAs in the regulation of macrophage polarization
Traditionally, the IRF/STAT signaling regulates either M1 or M2 polarization in macrophages (37-39). TLR ligands and IFN-γ stimulate STAT1 to activate IRF/STAT-mediated M1 responses, while IL-4/IL-10 stimulate STAT6 to induce IRF/STAT-mediated M2 responses (40). Given that miRNAs may target various transcription factors and adaptor proteins involved in IRF/STAT pathways, it is possible that these miRNAs could regulate either M1 or M2 polarization in macrophages. Indeed, as reviewed below, these miRNAs play distinctive roles in the regulation of macrophage polarization.
5.1 Controversial effects of miR-21 in modulating macrophage polarization
MiR-21 is one of the most highly studied miRNAs in mammalian cells due to its role in the pathophysiology of conditions such as solid tumors, cardiac injury and inflammation (41). However, there are contrasting views on the role of miR-21 in mediating macrophage polarization.
In a recent study performed by Caescu et al. (42), miR-21 was involved in the downstream effects of colony-stimulating factor 1 receptor (CSF-1R), which increased M2 macrophages and suppressed the M1 phenotype. Profiling of the mRNA targets of the CSF-1/miR-21 regulatory axis revealed that 80% of targets were pro-inflammatory molecules (42). Accordingly, knockdown of miR-21 resulted in the reduction of M2 phenotype genes, arginase 1, mannose receptor 1, IL-4Ra, and FIZZ (42). When miRNA-21 inhibitor was injected into mice, there was increased recruitment of inflammatory monocytes and increased response to LPS by peritoneal macrophages, thus signifying the ability of miR-21 to initiate M2 polarization (42). Moreover, these authors demonstrated that miR-21 suppressed levels of SIRPb1, a MEK/ERK1/2 pathway activator, further enhancing M2 responses (42).
In contrast to the findings by Caescu et al. (42), Wang et al. (43) reported that miR-21 promoted M1 polarization in peritoneal macrophages and inhibited M2 polarization. In miR-21-KO macrophages, expression levels of M1 markers (Tnfa, Il12p40, Il1b, and Il6) were lower while M2 markers (Il10, Arg1, Retnla, and Chi3l3) were increased remarkably compared to levels in wild type (WT) macrophages (43). Treatment of WT macrophages with prostaglandin E2 (PGE2), an endogenous lipid mediator produced in inflammation and infection, inhibited miR-21 expression (43). Further studies demonstrated that PGE2 decreased the expression of M1 markers and enhanced the expression of M2 markers (43). These data suggest that miR-21 acts as an endogenous brake to block PGE2-mediated generation of M2 macrophages. Mechanistically, there was an increase of STAT3 expression in miR-21-KO macrophages, which was further enhanced by PGE2 treatment (43). Luciferase assays also validated that miR-21 targeted STAT3 and thereby, promoted M1 polarization in macrophages (43).
At this moment, the reasons behind the conflicting effects of miR-21 on macrophage polarization remain unclear. However, there are some differences between the two studies described above. First, Caescu et al. (42) used bone marrow-derived macrophages, while Wang et al. (43) utilized peritoneal macrophages. Besides, Caescu et al. (42) elucidated effects on miR-21 expression in LPS-stimulated macrophages, while Wang et al. (43) investigated the role of miR-21 on macrophage polarization in miR-21-/- macrophages at basal expressions of different cytokines. Indeed, a recent study by Lu et al. (44) partially demonstrated such controversial effects of miR-21 on macrophage polarization by observing elevated miR-21 levels in both LPS-treated RAW 264.7 macrophages and IL-4 transgenic mouse lungs. Furthermore, according to Sheedy et al. (41), pri-miR-21 may be involved in the early inflammatory stage, while mature-miR-21 plays a role in the resolution phase. This observation may explain the contradictory observations in the findings described above. In the very early stage of inflammation, pri-miR-21 predominates and presents pro-inflammation activity and polarizes macrophages to the M1 phenotype. By contrast, at the resolution phase, mature-miR-21 regulates the anti-inflammatory responses and polarizes macrophages to the M2 phenotype (41). The authenticity of this theory, however, remains to be confirmed in future investigations.
5.2 MiRNAs in the regulation of M1 polarization
A recent study by Thulin et al. (45) showed that miR-9 enhanced M1 polarization by targeting peroxisome proliferator-activated receptor δ (PPARδ). The relative expression levels of miR-9 were increased, while PPARδ levels were decreased in human primary monocytes after LPS exposure (45). PPARδ is well known to regulate lipid and glucose homeostasis, as well as inflammation by activating the protein B cell lymphoma-6 (BCL-6), an anti-inflammatory transcriptional suppressor. Thus, increased expression of miR-9 resulted in suppression of PPARδ activity, thereby preventing the BCL-6-mediated anti-inflammatory effects. Consistently, transfection of anti-miR-9 in LPS-treated primary monocytes caused the increased expression of PPARδ (45). Hence, miR-9 may act to keep macrophages in the M1 polarized state.
Another miRNA reported to augment classical activation of macrophages is miR-127 (46). Inhibition of miR-127 expression by antagonist suppressed the expression of M1 signature genes and promoted transcription of M2 marker genes (46). Mechanistically, LPS-stimulated elevation of miR-127 inhibited the expression of Bcl6, which in turn suppressed the phosphatase Dusp1. Reduction of Dusp1 would lead to increased phosphorylation of JNK, which promoted the inflammatory response and M1 phenotype (46).
O'Connell et al. (47) recently observed that stimulation of macrophages with TLR2, TLR3, TLR4 and TLR9 ligands remarkably upregulated the expression of miR-155. Such an elevation of miR-155 was hampered by the knockdown of MyD88 and TRIF, two factors known to activate the NF-κB signaling. Likewise, the use of an JNK inhibitor diminished the upregulation of miR-155 and production of TNF-α, suggesting that miR-155-mediated activation of M1 polarization are due to induction of the JNK pathway (47). In addition, miR-155 can target the IL-13 receptor α1 (IL13Rα1), thereby inhibiting STAT6 activation and promoting M1 polarization (48). What's more, there were decreased levels of miR-155 in Akt2-/- macrophages, which coincided with increased levels of C/EBPβ, a bona fide target of miR-155 which has anti-inflammatory capacity (49). By contrast, higher expression of miR-155 was observed in Akt1-/- macrophages, in which the expression levels of M1 phenotype genes (TNF-α, IL-6 and iNOS), but not M2-associated genes (Arg1, Fiz1, and Ym1), were elevated (50). A luciferase reporter assay revealed that miR-155 interacted with the 3′UTR of suppressor of cytokine signaling 1 (SOCS1) (50). Therefore, these observations suggest that Akt signaling is also involved in the miR-155-mediated promotion of M1 phenotype through targeting C/EBPβ and SOCS1.
Finally, miR-125b has been demonstrated to positively regulate M1-phenotype in macrophages (51). Overexpression of miR-125b in macrophages was found to enhance responses of macrophages to M1 inducer IFN-γ through targeting of IRF4 (51). Accordingly, knockdown of IRF4 enhanced M1 activation and pro-inflammatory responses in macrophages (51).
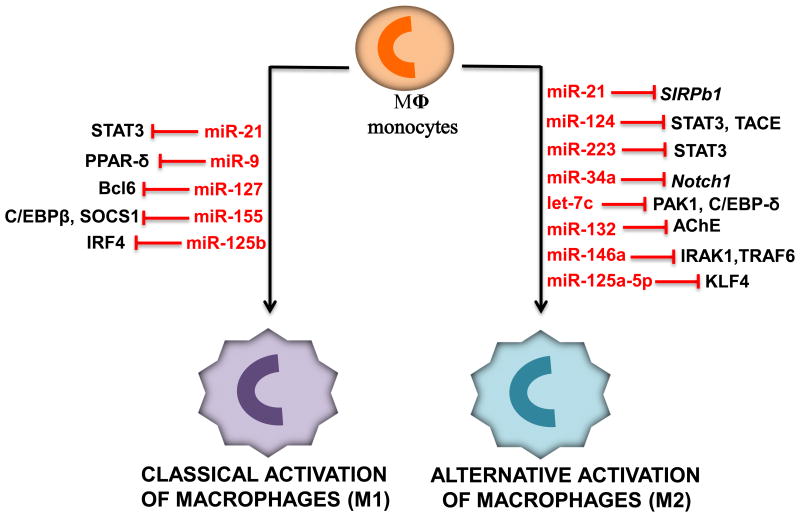
Taken together, miR-9, miR-127, miR-155 and miR-125b have been shown to promote classical activation of macrophages (M1) and pro-inflammatory responses through inhibiting various factors, as summarized in Figure 4 and Table 1.
Figure 4.
Diagram illustrates miRNAs that induce either classical activation or alternative activation of macrophages and their corresponding targets.
Table 1. miRNAs regulate M1 and M2 polarization through targeting various adaptor proteins and transcription factors.
| MiRNAs | Phenotype promoted | Targets | Function | Ref. |
|---|---|---|---|---|
|
| ||||
| miR-21 | M2 | SIRPb1 | Promotes M2 polarization, downstream of CSF-1R | [42] |
|
| ||||
| miR-21 | M1 | STAT3 | Prevents PGE2-Mediated M2 polarization | [43] |
|
| ||||
| miR-9 | M1 | PPAR-δ | Suppresses anti-inflammatory response | [45] |
|
| ||||
| miR-127 | M1 | Bcl6 | Promotes M1 phenotype through activation of JNK pathway | [46] |
|
| ||||
| miR-155 | M1 | ???? | Promotes pro-inflammatory responses via JNK pathway | [47] |
| C/EBPβ | Inhibits M2 polarization | [49] | ||
| SOCS1 | Promotes pro-inflammatory responses via P13K/Akt1 pathway | [50] | ||
|
| ||||
| miR-125b | M1 | IRF4 | Enhances pro-inflammatory responses | [51] |
|
| ||||
| miR-124 | M2 | STAT3, TACE | Inhibits production of pro-inflammatory cytokines | [52] |
|
| ||||
| miR-223 | M2 | STAT3 | Promotes anti-inflammatory response | [54] |
|
| ||||
| miR-34a | M2 | Notch1 | Inhibits production of pro-inflammatory cytokines | [56] |
|
| ||||
| Let-7c | M2 | PAK1 | Inhibits activation of NF-κB pathway | [57] |
| C/EBP-δ | Promotes M2 phenotype | [58] | ||
|
| ||||
| miR-132 | M2 | AChE | Promotes cholinergic anti-inflammatory response | [59] |
|
| ||||
| miR-146a | M2 | IRAK1,TRAF6 | Prevents activation of NF-κB | [61] |
|
| ||||
| miR-125a-5p | M2 | KLF4 | Promotes M2 polarization | [62] |
5.3 MiRNAs in the regulation of M2 polarization
Sun et al. (52) identified that miR-124 promoted cholinergic agonist-elicited anti-inflammatory effects. They observed that miR-124 was upregulated by cholinergic agonists in LPS-stimulated macrophages as well as in mice. Transfection of miR-124 mimics in RAW 264.7 cells resulted in a reduction of LPS-induced production of TNF-α and IL-6 (52). The authors further identified that miR-124 directly targeted STAT3 and TNF-α converting enzyme (TACE) to reduce IL-6 and TNF-α release, respectively (52). Similarly, Veremeyko et al. (53) observed that miR-124 expression was upregulated in both IL-4 and IL-13-treated macrophages, and knockdown of miR-124 suppressed the expression of M2 markers (i.e., CD206 and Ym1) and enhanced the expression of M1 markers (i.e., CD86, iNOS, TNF) in M2-polarized macrophages (53).
MiR-223 has also been shown to initiate M2 polarization in macrophages (54). MiR-223 is highly expressed in myeloid cells of the bone marrow, where it is mainly resident in the myeloid, granulocytic and monocytic compartments (55). Overexpression of miR-223 in RAW 264.7 macrophages inhibited LPS-stimulated release of IL-6 and IL-1β by targeting STAT3 (54). In addition, Jiang et al. (56) reported that miR-34a blocked pro-inflammatory responses in LPS-stimulated macrophages. MiR-34a levels were reduced in LPS-treated RAW 264.7 macrophages, but transfection of miR-34a mimics diminished pro-inflammatory responses, evidenced by lower levels of M1 cytokines TNF-α and IL-6 (56). Mechanistically, miR-34a targeted Notch1, which is needed for LPS-mediated production of pro-inflammatory cytokines in macrophages (56).
Let-7c has been reported to promote M2 phenotype through targeting of p21-activated kinase 1 (PAK1), a serine/threonine kinase that is upregulated in M1 polarized macrophages (57). This was evidenced by: 1) Levels of M1-associated genes such as TNF-α, IL-6, IL-12a, IL-12b, MCP-1, and Nos2 were decreased in LPS-treated PAK-/- BMDMs; 2) the introduction of let-7c mimics in LPS-treated RAW 264.7 macrophages diminished levels of PAK mRNA; and 3) enhancer of zeste homolog 2 (EZH2) suppressed levels of let-7c in M1 polarized macrophages, thereby preventing the inhibitory effects of let-7c on PAK1 (57). Furthermore, Banerjee et al. (58) observed that let-7c was highly expressed in M2 macrophages compared to M1 macrophages. Accordingly, overexpression of let-7c reduced the expression of M1 genes (i.e., IL-12 and iNOS), and increased levels of M2 marker (i.e., FR-β) in M1 macrophages via targeting C/EBP-δ (58). By contrast, knockdown of let-7c in M2 macrophages resulted in downregulation of M2 markers and upregulation of in M1 markers (58).
Additionally, miR-132 has been shown to elicit anti-inflammatory effects in alveolar macrophages (59). Treatment of rat alveolar macrophages with LPS resulted in an increase in miR-132 levels together with high levels of TNF-α, IL-1β, and IL-6 (59). Coinciding with the elevation of miR-132 was an initial increase in AChE protein levels but AChE levels declined at 24 h post-LPS treatment (59). Transfection of miR-132 mimics in ACh-pretreated macrophages repressed the pro-inflammatory responses upon LPS challenge through inhibition of the NF-κB and STAT3 pathways (59). Conversely, transfection of miR-132 inhibitor in ACh-pretreated macrophages enhanced production of TNF-α, IL-1β, and IL-6 (59). Similar to miR-132, miR-146a has the capacity to dampen the inflammatory response in alveolar macrophages (60). Transfection of peritoneal macrophages with miR-146a reduced the expression of M1-associated proteins (i.e., iNOS), and increased the expression of M2-associated genes after LPS treatment (60). Mechanistically, miR-146a may target NF-κB signaling mediators such as IRAK1 and TRAF6, thereby preventing pro-inflammatory responses (61). Lastly, miR-125a-5p targeted Kruppel-like factor 4 (KLF4), thereby inhibiting M1 polarization and promoting M2 polarization in bone marrow macrophages (62).
Collectively, miR-124, miR-223, miR-34a, let-7c, miR-132, miR-146a and miR-125a-5p have been identified thus far to promote anti-inflammatory responses and M2 polarization in macrophages, as summarized in Figure 4 and Table 1.
6. Therapeutic potential of miR-induced macrophage polarization in inflammation-related disease
The plasticity and diversity of macrophages in response to different pathophysiological conditions means that they could enhance or diminish progression of various inflammation-related disorders (63-66). Typically, classical activation (M1) is characterized by microbicidal and tumoricidal activity, whereas alternative activation (M2) is characterized with tumor progression and tissue remodeling (63-66). As such, miRNAs that regulate macrophage polarization may affect inflammation-related diseases such as sepsis, obesity, cancer and multiple sclerosis.
6.1 Macrophage polarization in sepsis
Sepsis is a severe inflammatory response syndrome that is a leading cause of death in the intensive care units of hospitals (67-70). Sepsis pathophysiology is complex, with the current consensus recognizing concurrent activation of pro-inflammatory and anti-inflammatory responses (71). However, it is generally believed that excessive production of pro-inflammatory cytokines in the initial stages of sepsis contributes to multiple organ damage observed in sepsis patients (67-71). This initial dominance of pro-inflammatory responses (systemic inflammatory response syndrome; SIRS) leads to a late resolution phase of higher anti-inflammatory cytokine production (compensatory anti-inflammatory response syndrome: CARS), which may counteract the earlier pro-inflammatory response (71). This resolution phase may lead to immunodepression and 'persistent inflammation – immunosuppression catabolism syndrome (PICS) which is characterized by reduction in lean body mass, poor wound healing, susceptibility to secondary infection and eventual death (71). Given the complicated balance between pro-inflammatory and anti-inflammatory responses, the role of macrophage polarization in sepsis is puzzled. Nevertheless, miRNAs that are involved in the macrophage polarization may contribute to the sepsis pathogenesis.
Indeed, numerous studies have implicated the beneficial effects of blocking TLR-NF-κB signaling in sepsis (72-78). Considering that TLR-NF-κB is essential for M1 activation (37), inhibiting those miRNAs that promote M1 polarization or activating the M2 polarization-associated miRNAs may have therapeutic potential in sepsis. For instance, serum concentrations of M2 phenotype-associated miRNAs (i.e., miR-146a and miR-223) were remarkably decreased in septic patients compared with SIRS patients and healthy controls (79). Accordingly, the beneficial effects of miR-146a in sepsis has been reported, where overexpression of miR-146a inhibited the expression of TLR4-NF-κB pathway proteins, IRAK1 and TRAF6, in cardiac monocytes, thereby blocking NF-κB activation and attenuating sepsis-induced cardiac dysfunction, inflammatory cell infiltration and inflammatory cytokine production (80). Although the mouse survival rate was increased, the authors did not investigate the effects of miR-146a overexpression on bacterial loads. Regarding the effects of circulating miR-223 in sepsis, Wang et al. (79) showed the correlation between reduced miR-223 levels and increased sepsis severity. However, a recent study by Benz et al. (81) suggested that serum levels of miR-223 did not predict sepsis prognosis or survival in patients with critical illness. Benz et al. (81) used samples from murine models of sepsis and human septic patients to conduct their studies. In the mouse model of cecal ligation and puncture (CLP) surgery, levels of circulating miR-223 remained unchanged at different time points after induction of sepsis compared to controls (81). In humans, serum levels of miR-223 were slightly lower in critically-ill patients compared with healthy controls (81). Importantly, no significant differences in miR-223 levels between septic and non-septic patients were evident, which is contradictory to findings by Wang et al. (79), where miR-223 was significantly decreased in septic patients compared to healthy individuals. The conflicting results between these two studies may be partially explained by differences in experimental procedures. The size and characteristics of the patient cohorts analyzed in these two studies may also influence the differences observed.
Currently, the definitive role of miRNA-induced macrophage polarization in sepsis is still unclear, although studies characterizing changes in miRNA expression in sepsis are extensive (72). As discussed above, changes in miR-146a and miR-223, which are confirmed modulators of macrophage polarization, seem to contribute to clinical outcomes in sepsis patients. Moving forward, miR-146a and miR-223 represent ideal miRNAs which could be utilized to investigate the role of miRNA-induced polarization in sepsis.
6.2 miR-induced macrophage polarization in other inflammation-related diseases
Polarized macrophages interact with various tissues and organs in diseases such as obesity (82-87), cancer (63-66) and multiple sclerosis (88,89), thus affecting pathophysiology of such disorders. MiRNAs that regulate activation or alternative activation of macrophages in these disease states may have potential for therapy. Overexpression of M2-related miRNAs (i.e., miR-223) (90, 91) and inhibition of M1-associated miRNAs (i.e., miR-33 and miR-155) (92, 93) have been shown to generate beneficial effects on obesity. Similarly, increased expression of pro-inflammatory miR-155 (88) and downregulation of anti-inflammatory miR-124 (89) has been linked to aggravation of multiple sclerosis. Finally, increased levels of miR-19a-3p (94), miR-16 (95), miR-155 (96,97) and miR-511-3p (98) promoted activation of tumor-associated macrophages (TAMs) and inhibition of tumor growth. On the other hand, overexpression of miR-146a (99), an M2 phenotype inducer, in TAMs was characterized with tumor progression.
7. Summary and future directions
Modulating inflammatory response has always been a hot topic for both basic and clinical research. Given that miRNAs epigenetically fine-tune hundreds of gene expression, there is increased attention on the regulatory roles of miRNAs in macrophage polarization. Especially, some miRNAs have been demonstrated to regulate the expression of various adaptor proteins and transcriptions factors, which are known to participate in macrophage polarization. Thus, alteration of such miRNA levels in macrophages should affect the switch between M1 and M2 phenotypes. As discussed above, some miRNAs (i.e., miR-9, miR-127, miR-155 and miR-125b) can promote M1 polarization, while other miRNAs (miR-124, miR-223, miR-34a, let-7c, miR-132, miR-146a and miR-125a-5p) can induce M2 polarization in both circulatory monocytes and tissue-resident macrophages. Future studies will be greatly needed to test whether such miRNAs can be utilized in clinical inflammation-related diseases. In addition, it would be significant to investigate whether dysregulation of polarization-associated miRNAs serves as biomarkers for prognosis and diagnosis of diseases.
Acknowledgments
Funding sources: The research in Dr. Guo-Chang Fan's lab is supported by NIH grants 2R01HL-087861 and R01 GM-112930.
References
- 1.Yang J, Zhang L, Yu C, Yang X-F, Wang H. Monocyte and macrophage differentiation: circulation inflammatory monocyte as biomarker for inflammatory diseases. Biomarker Research. 2014;2:1. doi: 10.1186/2050-7771-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Epelman S, Lavine KJ, Randolph GJ. Origin and functions of tissue macrophages. Immunity. 2014;41(1):21–35. doi: 10.1016/j.immuni.2014.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sica A, Mantovani A. Macrophage plasticity and polarization: in vivo veritas. J Clin Invest. 2012;122(3):787–95. doi: 10.1172/JCI59643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deiuliis JA. MicroRNAs as regulators of metabolic disease: pathophysiologic significance and emerging role as biomarkers and therapeutics. Int J Obes (Lond) 2015 doi: 10.1038/ijo.2015.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu G, Abraham E. MicroRNAs in immune response and macrophage polarization. Arterioscler Thromb Vasc Biol. 2013;33(2):170–7. doi: 10.1161/ATVBAHA.112.300068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466(7308):835–40. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang X, Huang W, Yang Y, Wang Y, Peng T, Chang J, Caldwell CC, Zingarelli B, Fan GC. Loss of duplexmiR-223 (5p and 3p) aggravates myocardial depression and mortality in polymicrobial sepsis. Biochim Biophys Acta. 2014;1842(5):701–11. doi: 10.1016/j.bbadis.2014.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang J, Li S, Li L, Li M, Guo C, Yao J, Mi S. Exosome and exosomal microRNA: trafficking, sorting, and function. Genomics Proteomics Bioinformatics. 2015;13(1):17–24. doi: 10.1016/j.gpb.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang X, Gu H, Qin D, Yang L, Huang W, Essandoh K, Wang Y, Caldwell CC, Peng T, Zingarelli B, Fan GC. Exosomal miR-223 Contributes to Mesenchymal Stem Cell-Elicited Cardioprotection in Polymicrobial Sepsis. Sci Rep. 2015;5:13721. doi: 10.1038/srep13721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alexander M, Hu R, Runtsch MC, Kagele DA, Mosbruger TL, Tolmachova T, Seabra MC, Round JL, Ward DM, O'Connell RM. Exosome-delivered microRNAs modulate the inflammatory response to endotoxin. Nat Commun. 2015;6:7321. doi: 10.1038/ncomms8321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Muñoz-Fernández MA, Fernández MA, Fresno M. Synergism between tumor necrosis factor-alpha and interferon-gamma on macrophage activation for the killing of intracellular Trypanosoma cruzi through a nitric oxide-dependent mechanism. Eur J Immunol. 1992;22(2):301–7. doi: 10.1002/eji.1830220203. [DOI] [PubMed] [Google Scholar]
- 13.Stout RD, Suttles J, Xu J, Grewal IS, Flavell RA. Impaired T cell-mediated macrophage activation in CD40 ligand-deficient mice. J Immunol. 1996;156(1):8–11. [PubMed] [Google Scholar]
- 14.Bekker LG, Freeman S, Murray PJ, Ryffel B, Kaplan G. TNF-alpha controls intracellular mycobacterial growth by both inducible nitric oxide synthase-dependent and inducible nitric oxide synthase-independent pathways. J Immunol. 2001;166(11):6728–34. doi: 10.4049/jimmunol.166.11.6728. [DOI] [PubMed] [Google Scholar]
- 15.Ehlers S, Kutsch S, Ehlers EM, Benini J, Pfeffer K. Lethal granuloma disintegration in mycobacteria-infected TNFRp55-/- mice is dependent on T cells and IL-12. J Immunol. 2000;165(1):483–92. doi: 10.4049/jimmunol.165.1.483. [DOI] [PubMed] [Google Scholar]
- 16.Chávez-Galán L, Olleros ML, Vesin D, Garcia I. Much More than M1 and M2 Macrophages, There are also CD169(+) and TCR(+) Macrophages. Front Immunol. 2015;6:263. doi: 10.3389/fimmu.2015.00263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beutler B, Rietschel ET. Innate immune sensing and its roots: the story of endotoxin. Nat Rev Immunol. 2003;3(2):169–76. doi: 10.1038/nri1004. [DOI] [PubMed] [Google Scholar]
- 18.Wicks IP, Roberts AW. Targeting GM-CSF in inflammatory diseases. Nat Rev Rheumatol. 2015 doi: 10.1038/nrrheum.2015.161. [DOI] [PubMed] [Google Scholar]
- 19.Hao NB, Lü MH, Fan YH, Cao YL, Zhang ZR, Yang SM. Macrophages in tumor microenvironments and the progression of tumors. Clin Dev Immunol. 2012;2012:948098. doi: 10.1155/2012/948098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mantovani A, Sozzani S, Locati M, Allavena P, Sica A. Macrophage polarization: tumor-associated macrophages as a paradigm for polarized M2 mononuclear phagocytes. Trends Immunol. 2002;23(11):549–55. doi: 10.1016/s1471-4906(02)02302-5. [DOI] [PubMed] [Google Scholar]
- 21.Gordon S. Alternative activation of macrophages. Nat Rev Immunol. 2003;3(1):23–35. doi: 10.1038/nri978. [DOI] [PubMed] [Google Scholar]
- 22.Auphan N, DiDonato JA, Rosette C, Helmberg A, Karin M. Immunosuppression by glucocorticoids: inhibition of NF-kappa B activity through induction of I kappa B synthesis. Science. 1995;270(5234):286–90. doi: 10.1126/science.270.5234.286. [DOI] [PubMed] [Google Scholar]
- 23.Herrlich P. Cross-talk between glucocorticoid receptor and AP-1. Oncogene. 2001;20(19):2465–75. doi: 10.1038/sj.onc.1204388. [DOI] [PubMed] [Google Scholar]
- 24.Bhattacharyya S, Brown DE, Brewer JA, Vogt SK, Muglia LJ. Macrophage glucocorticoid receptors regulate Toll-like receptor 4–mediated inflammatory responses by selective inhibition of p38 MAP kinase. Blood. 2007;109(10):4313–4319. doi: 10.1182/blood-2006-10-048215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liden J, Rafter I, Truss M, Gustafsson JA, Okret S. Glucocorticoid effects on NF-kappaB binding in the transcription of the ICAM-1 gene. Biochem Biophys Res Commun. 2000;273(3):1008–14. doi: 10.1006/bbrc.2000.3079. [DOI] [PubMed] [Google Scholar]
- 26.Penton-Rol G, Cota M, Polentarutti N, Luini W, Bernasconi S, Borsatti A, Sica A, LaRosa GJ, Sozzani S, Poli G, Mantovani A. Up-regulation of CCR2 chemokine receptor expression and increased susceptibility to the multitropic HIV strain 89.6 in monocytes exposed to glucocorticoid hormones. J Immunol. 1999;163(6):3524–9. [PubMed] [Google Scholar]
- 27.Wang Q, Ni H, Lan L, Wei X, Xiang R, Wang Y. Fra-1 protooncogene regulates IL-6 expression in macrophages and promotes the generation of M2d macrophages. Cell Res. 2010;20(6):701–12. doi: 10.1038/cr.2010.52. [DOI] [PubMed] [Google Scholar]
- 28.Duluc D, Delneste Y, Tan F, Moles MP, Grimaud L, Lenoir J, Preisser L, Anegon I, Catala L, Ifrah N, Descamps P, Gamelin E, Gascan H, Hebbar M, Jeannin P. Tumor-associated leukemia inhibitory factor and IL-6 skew monocyte differentiation into tumor-associated macrophage-like cells. Blood. 2007;110(13):4319–30. doi: 10.1182/blood-2007-02-072587. [DOI] [PubMed] [Google Scholar]
- 29.Chanmee T, Ontong P, Konno K, Itano N. Tumor-Associated Macrophages as Major Players in the Tumor Microenvironment. Cancers. 2014;6(3):1670–1690. doi: 10.3390/cancers6031670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jaguin M, Houlbert N, Fardel O, Lecureur V. Polarization profiles of human M-CSF-generated macrophages and comparison of M1-markers in classically activated macrophages from GM-CSF and M-CSF origin. Cell Immunol. 2013;281(1):51–61. doi: 10.1016/j.cellimm.2013.01.010. [DOI] [PubMed] [Google Scholar]
- 31.Hattermann K, Sebens S, Helm O, Schmitt AD, Mentlein R, Mehdorn HM, Held-Feindt J. Chemokine expression profile of freshly isolated human glioblastoma-associated macrophages/microglia. Oncol Rep. 2014;32(1):270–6. doi: 10.3892/or.2014.3214. [DOI] [PubMed] [Google Scholar]
- 32.Zhang Y, Zhang M, Zhong M, Suo Q, Lv K. Expression profiles of miRNAs in polarized macrophages. Int J Mol Med. 2013;31(4):797–802. doi: 10.3892/ijmm.2013.1260. [DOI] [PubMed] [Google Scholar]
- 33.Cobos Jiménez V, Bradley EJ, Willemsen AM, van Kampen AH, Baas F, Kootstra NA. Next-generation sequencing of microRNAs uncovers expression signatures in polarized macrophages. Physiol Genomics. 2014;46(3):91–103. doi: 10.1152/physiolgenomics.00140.2013. [DOI] [PubMed] [Google Scholar]
- 34.Murray PJ, Allen JE, Biswas SK, Fisher EA, Gilroy DW, Goerdt S, Gordon S, Hamilton JA, Ivashkiv LB, Lawrence T, Locati M, Mantovani A, Martinez FO, Mege JL, Mosser DM, Natoli G, Saeij JP, Schultze JL, Shirey KA, Sica A, Suttles J, Udalova I, van Ginderachter JA, Vogel SN, Wynn TA. Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity. 2014;41(1):14–20. doi: 10.1016/j.immuni.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Davies LC, Taylor PR. Tissue-resident macrophages: then and now. Immunology. 2015;144(4):541–8. doi: 10.1111/imm.12451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Graff JW, Dickson AM, Clay G, McCaffrey AP, Wilson ME. Identifying functional microRNAs in macrophages with polarized phenotypes. J Biol Chem. 2012;287(26):21816–25. doi: 10.1074/jbc.M111.327031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lawrence T, Natoli G. Transcriptional regulation of macrophage polarization: enabling diversity with identity. Nat Rev Immunol. 2011;11(11):750–61. doi: 10.1038/nri3088. [DOI] [PubMed] [Google Scholar]
- 38.Günthner R, Anders HJ. Interferon-regulatory factors determine macrophage phenotype polarization. Mediators Inflamm. 2013;2013:731023. doi: 10.1155/2013/731023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sica A, Mantovani A. Macrophage plasticity and polarization: in vivo veritas. J Clin Invest. 2012;122(3):787–95. doi: 10.1172/JCI59643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang N, Liang H, Zen K. Molecular mechanisms that influence the macrophage m1-m2 polarization balance. Front Immunol. 2014;5:614. doi: 10.3389/fimmu.2014.00614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sheedy FJ. Turning 21: Induction of miR-21 as a Key Switch in the Inflammatory Response. Front Immunol. 2015;6:19. doi: 10.3389/fimmu.2015.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Caescu CI, Guo X, Tesfa L, Bhagat TD, Verma A, Zheng D, Stanley ER. Colony stimulating factor-1 receptor signaling networks inhibit mouse macrophage inflammatory responses by induction of microRNA-21. Blood. 2015;125(8):e1–13. doi: 10.1182/blood-2014-10-608000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang Z, Brandt S, Medeiros A, Wang S, Wu H, Dent A, Serezani CH. MicroRNA 21 is a homeostatic regulator of macrophage polarization and prevents prostaglandin E2-mediated M2 generation. PLoS One. 2015;10(2):e0115855. doi: 10.1371/journal.pone.0115855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lu TX, Munitz A, Rothenberg ME. MicroRNA-21 is up-regulated in allergic airway inflammation and regulates IL-12p35 expression. J Immunol. 2009;182(8):4994–5002. doi: 10.4049/jimmunol.0803560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thulin P, Wei T, Werngren O, Cheung L, Fisher RM, Grandér D, Corcoran M, Ehrenborg E. MicroRNA-9 regulates the expression of peroxisome proliferator-activated receptor δ in human monocytes during the inflammatory response. Int J Mol Med. 2013;31(5):1003–10. doi: 10.3892/ijmm.2013.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ying H, Kang Y, Zhang H, Zhao D, Xia J, Lu Z, Wang H, Xu F, Shi L. MiR-127 modulates macrophage polarization and promotes lung inflammation and injury by activating the JNK pathway. J Immunol. 2015;194(3):1239–51. doi: 10.4049/jimmunol.1402088. [DOI] [PubMed] [Google Scholar]
- 47.O'Connell RM, Taganov KD, Boldin MP, Cheng G, Baltimore D. MicroRNA-155 is induced during the macrophage inflammatory response. Proc Natl Acad Sci USA. 2007;104(5):1604–9. doi: 10.1073/pnas.0610731104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martinez-Nunez RT, Louafi F, Sanchez-Elsner T. The interleukin 13 (IL-13) pathway in human macrophages is modulated by microRNA-155 via direct targeting of interleukin 13 receptor alpha1 (IL13Ralpha1) J Biol Chem. 2011;286(3):1786–94. doi: 10.1074/jbc.M110.169367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Arranz A, Doxaki C, Vergadi E, Martinez de la Torre Y, Vaporidi K, Lagoudaki ED, Ieronymaki E, Androulidaki A, Venihaki M, Margioris AN, Stathopoulos EN, Tsichlis PN, Tsatsanis C. Akt1 and Akt2 protein kinases differentially contribute to macrophage polarization. Proc Natl Acad Sci USA. 2012;109(24):9517–22. doi: 10.1073/pnas.1119038109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xu F, Kang Y, Zhang H, Piao Z, Yin H, Diao R, Xia J, Shi L. Akt1-mediated regulation of macrophage polarization in a murine model of Staphylococcus aureus pulmonary infection. J Infect Dis. 2013;208(3):528–38. doi: 10.1093/infdis/jit177. [DOI] [PubMed] [Google Scholar]
- 51.Chaudhuri AA, So AY, Sinha N, Gibson WS, Taganov KD, O'Connell RM, Baltimore D. MicroRNA-125b potentiates macrophage activation. J Immunol. 2011;187(10):5062–8. doi: 10.4049/jimmunol.1102001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sun Y, Li Q, Gui H, Xu DP, Yang YL, Su DF, Liu X. MicroRNA-124 mediates the cholinergic anti-inflammatory action through inhibiting the production of pro-inflammatory cytokines. Cell Res. 2013;23(11):1270–83. doi: 10.1038/cr.2013.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Veremeyko T, Siddiqui S, Sotnikov I, Yung A, Ponomarev ED. IL-4/IL-13-dependent and independent expression of miR-124 and its contribution to M2 phenotype of monocytic cells in normal conditions and during allergic inflammation. PLoS One. 2013;8(12):e81774. doi: 10.1371/journal.pone.0081774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen Q, Wang H, Liu Y, Song Y, Lai L, Han Q, Cao X, Wang Q. Inducible microRNA-223 down-regulation promotes TLR-triggered IL-6 and IL-1β production in macrophages by targeting STAT3. PLoS One. 2012;7(8):e42971. doi: 10.1371/journal.pone.0042971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Haneklaus M, Gerlic M, O'Neill LA, Masters SL. miR-223: infection, inflammation and cancer. J Intern Med. 2013;274(3):215–26. doi: 10.1111/joim.12099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jiang P, Liu R, Zheng Y, Liu X, Chang L, Xiong S, Chu Y. MiR-34a inhibits lipopolysaccharide-induced inflammatory response through targeting Notch1 in murine macrophages. Exp Cell Res. 2012;318(10):1175–84. doi: 10.1016/j.yexcr.2012.03.018. [DOI] [PubMed] [Google Scholar]
- 57.Zhang W, Liu H, Liu W, Liu Y, Xu J. Polycomb-mediated loss of microRNA let-7c determines inflammatory macrophage polarization via PAK1-dependent NF-κB pathway. Cell Death Differ. 2015;22(2):287–97. doi: 10.1038/cdd.2014.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Banerjee S, Xie N, Cui H, Tan Z, Yang S, Icyuz M, Abraham E, Liu G. MicroRNA let-7c regulates macrophage polarization. J Immunol. 2013;190(12):6542–9. doi: 10.4049/jimmunol.1202496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu F, Li Y, Jiang R, Nie C, Zeng Z, Zhao N, Huang C, Shao Q, Ding C, Qing C, Xia L, Zeng E, Qian K. miR-132 inhibits lipopolysaccharide-induced inflammation in alveolar macrophages by the cholinergic anti-inflammatory pathway. Exp Lung Res. 2015;41(5):261–9. doi: 10.3109/01902148.2015.1004206. [DOI] [PubMed] [Google Scholar]
- 60.Vergadi E, Vaporidi K, Theodorakis EE, Doxaki C, Lagoudaki E, Ieronymaki E, Alexaki VI, Helms M, Kondili E, Soennichsen B, Stathopoulos EN, Margioris AN, Georgopoulos D, Tsatsanis C. Akt2 deficiency protects from acute lung injury via alternative macrophage activation and miR-146a induction in mice. J Immunol. 2014;192(1):394–406. doi: 10.4049/jimmunol.1300959. [DOI] [PubMed] [Google Scholar]
- 61.Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci USA. 2006;103(33):12481–6. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Banerjee S, Cui H, Xie N, Tan Z, Yang S, Icyuz M, Thannickal VJ, Abraham E, Liu G. miR-125a-5p regulates differential activation of macrophages and inflammation. J Biol Chem. 2013;288(49):35428–36. doi: 10.1074/jbc.M112.426866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Labonte AC, Tosello-Trampont A-C, Hahn YS. The Role of Macrophage Polarization in Infectious and Inflammatory Diseases. Molecules and Cells. 2014;37(4):275–285. doi: 10.14348/molcells.2014.2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bashir S, Sharma Y, Elahi A, Khan F. Macrophage polarization: the link between inflammation and related diseases. Inflamm Res. 2015 doi: 10.1007/s00011-015-0874-1. [DOI] [PubMed] [Google Scholar]
- 65.Biswas SK, Chittezhath M, Shalova IN, Lim JY. Macrophage polarization and plasticity in health and disease. Immunol Res. 2012;53(1-3):11–24. doi: 10.1007/s12026-012-8291-9. [DOI] [PubMed] [Google Scholar]
- 66.Liu YC, Zou XB, Chai YF, Yao YM. Macrophage polarization in inflammatory diseases. Int J Biol Sci. 2014;10(5):520–9. doi: 10.7150/ijbs.8879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hawiger J, Veach RA, Zienkiewicz J. New paradigms in sepsis: from prevention to protection of failing microcirculation. J Thromb Haemost. 2015;13(10):1743–56. doi: 10.1111/jth.13061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mayr FB, Yende S, Angus DC. Epidemiology of severe sepsis. Virulence. 2014;5(1):4–11. doi: 10.4161/viru.27372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Martin GS. Sepsis, severe sepsis and septic shock: changes in incidence, pathogens and outcomes. Expert review of anti-infective therapy. 2012;10(6):701–706. doi: 10.1586/eri.12.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Melamed A, Sorvillo FJ. The burden of sepsis-associated mortality in the United States from 1999 to 2005: an analysis of multiple-cause-of-death data. Crit Care. 2009;13(1):R28. doi: 10.1186/cc7733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gentile LF, Cuenca AG, Efron PA, Ang D, Bihorac A, McKinley BA, Moldawer LL, Moore FA. Persistent inflammation and immunosuppression: a common syndrome and new horizon for surgical intensive care. J Trauma Acute Care Surg. 2012;72(6):1491–501. doi: 10.1097/TA.0b013e318256e000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Essandoh K, Fan GC. Role of extracellular and intracellular microRNAs in sepsis. Biochim Biophys Acta. 2014;1842(11):2155–2162. doi: 10.1016/j.bbadis.2014.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tsujimoto H, Ono S, Efron PA, Scumpia PO, Moldawer LL, Mochizuki H. Role of Toll-like receptors in the development of sepsis. Shock. 2008;29(3):315–21. doi: 10.1097/SHK.0b013e318157ee55. [DOI] [PubMed] [Google Scholar]
- 74.Cristofaro P, Opal SM. The Toll-like receptors and their role in septic shock. Expert Opin Ther Targets. 2003;7(5):603–12. doi: 10.1517/14728222.7.5.603. [DOI] [PubMed] [Google Scholar]
- 75.Savva A, Roger T. Targeting toll-like receptors: promising therapeutic strategies for the management of sepsis-associated pathology and infectious diseases. Front Immunol. 2013;4:387. doi: 10.3389/fimmu.2013.00387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Weighardt H, Holzmann B. Role of Toll-like receptor responses for sepsis pathogenesis. Immunobiology. 2007;212(9-10):715–22. doi: 10.1016/j.imbio.2007.09.010. [DOI] [PubMed] [Google Scholar]
- 77.Foley NM, Wang J, Redmond HP, Wang JH. Current knowledge and future directions of TLR and NOD signaling in sepsis. Military Medical Research. 2015;2:1. doi: 10.1186/s40779-014-0029-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Salomão R, Martins PS, Brunialti MK, Fernandes Mda L, Martos LS, Mendes ME, Gomes NE, Rigato O. TLR signaling pathway in patients with sepsis. Shock. 2008;30(Suppl 1):73–7. doi: 10.1097/SHK.0b013e318181af2a. [DOI] [PubMed] [Google Scholar]
- 79.Wang JF, Yu ML, Yu G, Bian JJ, Deng XM, Wan XJ, Zhu KM. Serum miR-146a and miR-223 as potential new biomarkers for sepsis. Biochem Biophys Res Commun. 2010;394(1):184–8. doi: 10.1016/j.bbrc.2010.02.145. [DOI] [PubMed] [Google Scholar]
- 80.Gao M, Wang X, Zhang X, Ha T, Ma H, Liu L, Kalbfleisch JH, Gao X, Kao RL, Williams DL, Li C. Attenuation of Cardiac Dysfunction in Polymicrobial Sepsis by MicroRNA-146a Is Mediated via Targeting of IRAK1 and TRAF6 Expression. J Immunol. 2015;195(2):672–82. doi: 10.4049/jimmunol.1403155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Benz F, Tacke F, Luedde M, Trautwein C, Luedde T, Koch A, Roderburg C. Circulating microRNA-223 serum levels do not predict sepsis or survival in patients with critical illness. Dis Markers. 2015;2015:384208. doi: 10.1155/2015/384208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Arner P, Kulyté A. MicroRNA regulatory networks in human adipose tissue and obesity. Nat Rev Endocrinol. 2015;11(5):276–88. doi: 10.1038/nrendo.2015.25. [DOI] [PubMed] [Google Scholar]
- 83.Monteiro R, Azevedo I. Chronic Inflammation in Obesity and the Metabolic Syndrome. Mediators of Inflammation. 2010;2010:289645. doi: 10.1155/2010/289645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Xu H, Barnes GT, Yang Q, Tan G, Yang D, Chou CJ, Sole J, Nichols A, Ross JS, Tartaglia LA, Chen H. Chronic inflammation in fat plays a crucial role in the development of obesity-related insulin resistance. J Clin Invest. 2003;112(12):1821–30. doi: 10.1172/JCI19451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gregor MF, Hotamisligil GS. Inflammatory mechanisms in obesity. Annu Rev Immunol. 2011;29:415–45. doi: 10.1146/annurev-immunol-031210-101322. [DOI] [PubMed] [Google Scholar]
- 86.Nishimura S, Manabe I, Nagai R. Adipose tissue inflammation in obesity and metabolic syndrome. Discov Med. 2009;8(41):55–60. [PubMed] [Google Scholar]
- 87.Bastard JP, Maachi M, Lagathu C, Kim MJ, Caron M, Vidal H, Capeau J, Feve B. Recent advances in the relationship between obesity, inflammation, and insulin resistance. Eur Cytokine Netw. 2006;17(1):4–12. [PubMed] [Google Scholar]
- 88.Moore CS, Rao VT, Durafourt BA, Bedell BJ, Ludwin SK, Bar-Or A, Antel JP. miR-155 as a multiple sclerosis-relevant regulator of myeloid cell polarization. Ann Neurol. 2013;74(5):709–20. doi: 10.1002/ana.23967. [DOI] [PubMed] [Google Scholar]
- 89.Ponomarev ED, Veremeyko T, Barteneva N, Krichevsky AM, Weiner HL. MicroRNA-124 promotes microglia quiescence and suppresses EAE by deactivating macrophages via the C/EBP-α-PU. 1 pathway Nat Med. 2011;17(1):64–70. doi: 10.1038/nm.2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ying W, Tseng A, Chang RC, Morin A, Brehm T, Triff K, Nair V, Zhuang G, Song H, Kanameni S, Wang H, Golding MC, Bazer FW, Chapkin RS, Safe S, Zhou B. MicroRNA-223 is a crucial mediator of PPARγ-regulated alternative macrophage activation. J Clin Invest. 2015;125(11):4149–4159. doi: 10.1172/JCI81656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhuang G, Meng C, Guo X, Cheruku PS, Shi L, Xu H, Li H, Wang G, Evans AR, Safe S, Wu C, Zhou B. A novel regulator of macrophage activation: miR-223 in obesity-associated adipose tissue inflammation. Circulation. 2012;125(23):2892–903. doi: 10.1161/CIRCULATIONAHA.111.087817. [DOI] [PubMed] [Google Scholar]
- 92.Karunakaran D, Richards L, Geoffrion M, Barrette D, Gotfrit RJ, Harper ME, Rayner KJ. Therapeutic Inhibition of miR-33 Promotes Fatty Acid Oxidation but Does Not Ameliorate Metabolic Dysfunction in Diet-Induced Obesity. Arterioscler Thromb Vasc Biol. 2015;35(12):2536–43. doi: 10.1161/ATVBAHA.115.306404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yang Y, Yang L, Liang X, Zhu G. MicroRNA-155 Promotes Atherosclerosis Inflammation via Targeting SOCS1. Cell Physiol Biochem. 2015;36(4):1371–81. doi: 10.1159/000430303. [DOI] [PubMed] [Google Scholar]
- 94.Yang J, Zhang Z, Chen C, Liu Y, Si Q, Chuang TH, Li N, Gomez-Cabrero A, Reisfeld RA, Xiang R, Luo Y. MicroRNA-19a-3p inhibits breast cancer progression and metastasis by inducing macrophage polarization through downregulated expression of Fra-1 proto-oncogene. Oncogene. 2014;33(23):3014–23. doi: 10.1038/onc.2013.258. [DOI] [PubMed] [Google Scholar]
- 95.Jang JY, Lee JK, Jeon YK, Kim CW. Exosome derived from epigallocatechin gallate treated breast cancer cells suppresses tumor growth by inhibiting tumor-associated macrophage infiltration and M2 polarization. BMC Cancer. 2013;13:421. doi: 10.1186/1471-2407-13-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Cai X, Yin Y, Li N, Zhu D, Zhang J, Zhang CY, Zen K. Re-polarization of tumor-associated macrophages to pro-inflammatory M1 macrophages by microRNA-155. J Mol Cell Biol. 2012;4(5):341–3. doi: 10.1093/jmcb/mjs044. [DOI] [PubMed] [Google Scholar]
- 97.Yu F, Jia X, Du F, Wang J, Wang Y, Ai W, Fan D. miR-155-deficient bone marrow promotes tumor metastasis. Mol Cancer Res. 2013;11(8):923–36. doi: 10.1158/1541-7786.MCR-12-0686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Squadrito ML, Pucci F, Magri L, Moi D, Gilfillan GD, Ranghetti A, Casazza A, Mazzone M, Lyle R, Naldini L, De Palma M. miR-511-3p modulates genetic programs of tumor-associated macrophages. Cell Rep. 2012;1(2):141–54. doi: 10.1016/j.celrep.2011.12.005. [DOI] [PubMed] [Google Scholar]
- 99.Perske C, Lahat N, Sheffy Levin S, Bitterman H, Hemmerlein B, Rahat MA. Loss of inducible nitric oxide synthase expression in the mouse renal cell carcinoma cell line RENCA is mediated by microRNA miR-146a. Am J Pathol. 2010;177(4):2046–54. doi: 10.2353/ajpath.2010.091111. [DOI] [PMC free article] [PubMed] [Google Scholar]