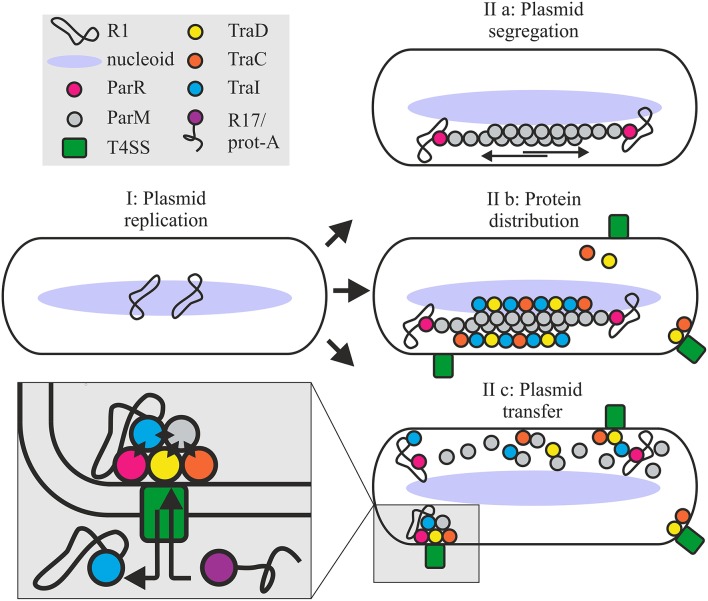
Figure 8.
Roles for the ParMRC system in plasmid propagation. Newly replicated plasmids are located at midcell (I). ParR bound to centromere parC captures and protects the end of a growing ParM filament. Two antiparallel ParM filaments create biopolar spindles which elongate and actively segregate plasmids to opposite ends of the dividing cell (IIa). Affinity of Tra proteins for ParM concentrates these along the filament longitudinal axis promoting assembly of the T4SS (IIb). Once spindles deliver the plasmid to the cell poles ParM filaments depolymerize releasing the DNA and protein cargo. ParM and ParR proteins become integrated at the interface of relaxosome, T4CP and channel ATPase TraC (IIc) via multiple protein-protein interactions as shown by black diamonds in the expanded view. Mixed assembly of Tra proteins, Par proteins and relaxosome bring the T4SS components and or the extracellular pilus in a conformation ideally primed for conjugative DNA transfer. This optimized conformational state supports robust biofilm formation by the bacterium and renders the T4SS competent to take up the protein A-R17 RNA complex during phage infection.