Figure 7.
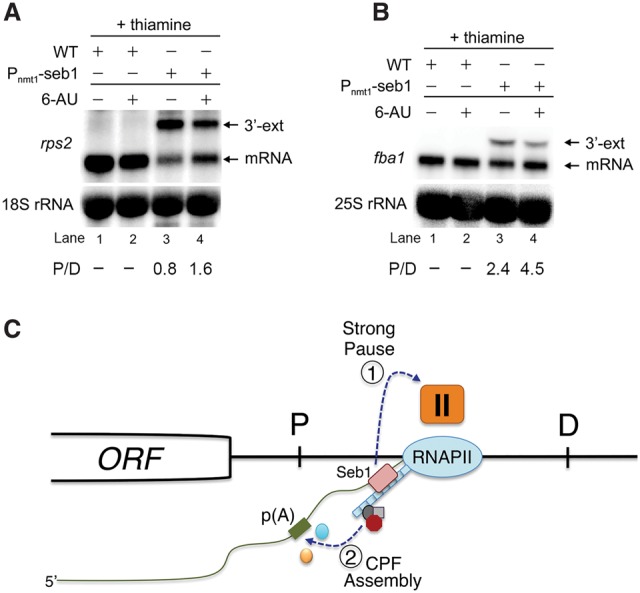
Transcription kinetics contributes to Seb1-dependent poly(A) site selection. (A,B) Northern blot analysis of total RNA prepared from wild-type (lanes 1,2) and Seb1-depleted (lanes 3,4) cells that were treated (lanes 2,4) or not treated (lanes 1,3) with 6-AU. Blots were probed for rps2 (A) and fba1 (B) mRNAs. Ratios of proximal (P) relative to distal (D) mRNA isoforms are indicated (average from two independent experiments). (C) Model for Seb1-dependent poly(A) site selection. The passage of RNAPII through a poly(A) site is thought to induce a change in the kinetics of transcription elongation, including pausing of the RNAPII complex (Nag et al. 2006; Grosso et al. 2012; Davidson et al. 2014; Fusby et al. 2015; Nojima et al. 2015). We propose that the cooperative binding of Seb1 to the RNAPII CTD and to RNA motifs clustered downstream from poly(A) sites positively contributes to RNAPII pausing (1), thereby promoting poly(A) site recognition and assembly of a cleavage-competent cleavage/poly(A) (CPF) complex (2). In the absence of Seb1, RNAPII pausing is leaky, increasing the frequency of RNAPII complexes that reach distal (D) poly(A) sites.
