Figure 4.
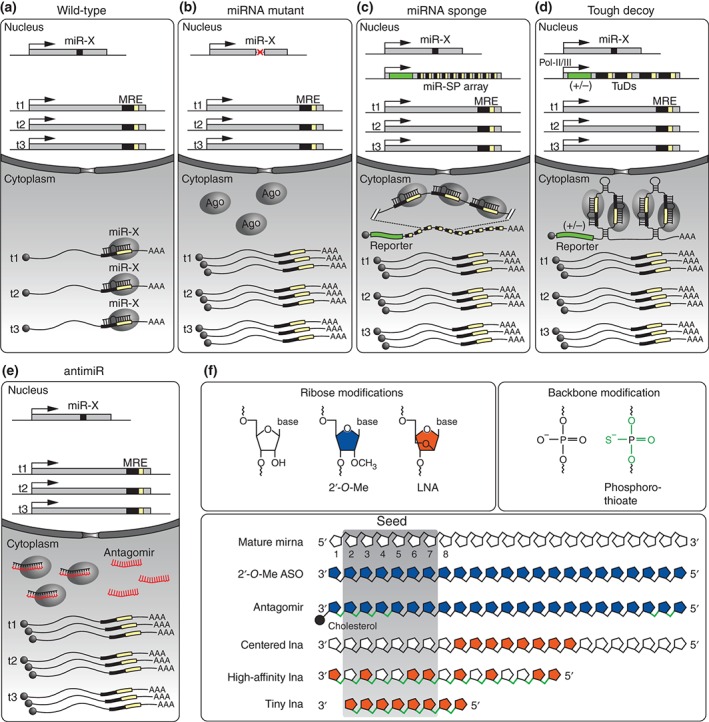
miRNA loss‐of‐function approaches. (a) Under normal homeostasis, endogenous miRNA‐activity results in target repression. (b) Ablation of the genomic miRNA locus enables generation of a null miRNA mutant allele resulting in a global de‐repression of all its targets. (c) miRNA sponge (miR‐SP) competitive inhibitors are genetically encoded into the 3′UTR of a reporter genes and expressed from Pol II promoters. When deployed to in vivo they sequester a cognate miRNAs resulting in de‐repression of its endogenous targets. (d) Tough decoys (TuD), are also genetically encoded miRNA competitive inhibitors with enhanced RNA secondary structure for improved targeting and stability. (e) Modified synthetic antimiR oligonucleotides (such as antagomiRs) can be transfected into cells to sequester miRNAs away from their targets. (f) AntimiRs frequently contain chemical modifications which enhance their cellular stability and increase their miRNA binding affinity. Common ribose modifications include 2′‐O‐methyl nucleotides or locked nucleic acid (LNA) incorporations for enhanced affinity. The phosphate backbone can also carry a phosphorothioate modification for increased resistance to RNase degradation. Antagomirs are coupled to cholesterol to aid cellular uptake. LNA nucleotides have been incorporated in the center of long ASOs, spaced throughout the entire length of the ASO. In contrast, tiny LNAs are entirely LNA‐based short 8 bp ASOs complementary to the miRNA seed sequence. MRE, miRNA response element; Pol‐II/III, RNA Polymerase II/III promoters; t1,2,3, miRNA targets; AAA, poly‐A tail; miR‐SP, miRNA sponge; TuD, Tough decoy; ASO, antisense oligonucleotides; yellow box in MRE, seed match region.
