Figure 1.
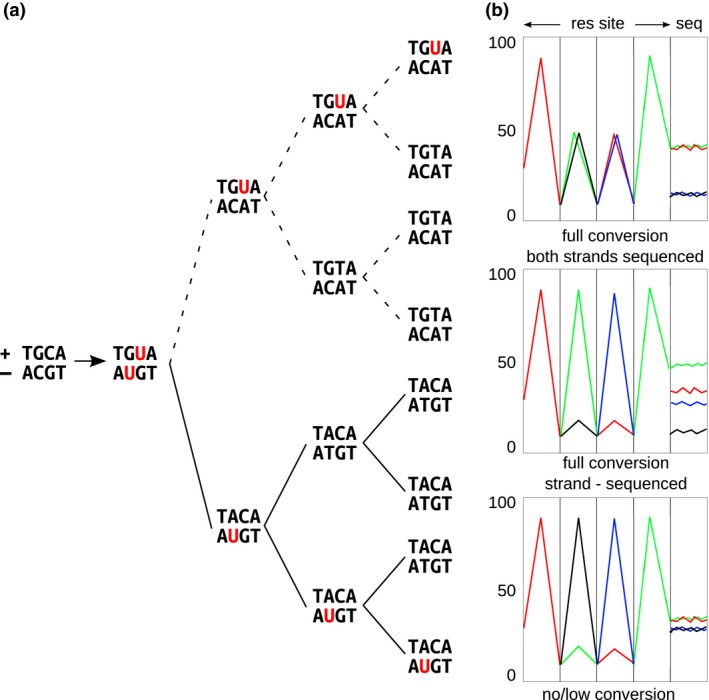
Sketch of directional bisulfite conversion used in bsRADseq: (a) Nonmethylated cytosines in the overhanging portion of the restriction site left by SbfI are converted by bisulfite treatment on both strands, ‘+’ and ‘−‘ (red). The converted ‘−‘ strand serves as template during RADseq directional amplification. Sequencing of the complementary to converted ‘−‘ strand is then carried out. (b) Three of the possible sequencing products, showing the bases of the restriction site (res site) and the expected average base composition in the remaining positions over all reads (seq): full bisulfite conversion of cytosines and sequencing of both strands (upper panel); full bisulfite conversion of cytosines but sequencing of the ‘−‘ strand only (middle panel); no/low bisulfite conversion of cytosines and sequencing of both strands (lower panel). The middle panel outcome was observed in our bisulfite conversion and sequencing experiments. Proportion of reads containing the specific nucleotide is shown on the y‐axis. Standard color code (thymine: red, cytosine: blue, adenine: green, guanine: black) is used.
