Figure 7.
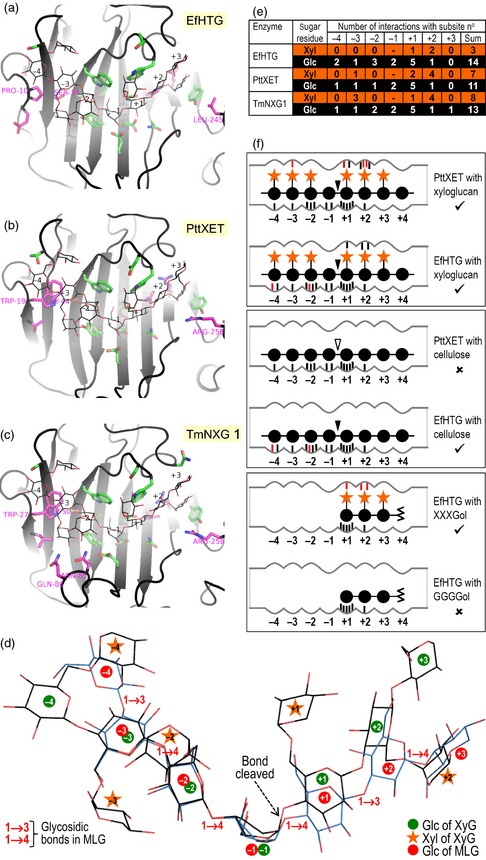
Modelled interaction of substrates with the active site in HTG and classical XTHs.
(a–c) Substrate binding pocket of EfHTG and two XTHs (PttXET and TmNXG1). Grey ribbon, protein backbone; line drawing, XXXGXXG; sticks with green carbons, side chains lining the active‐site cleft; magenta carbons, residues that are functionally non‐conserved between HTG and both PttXET and TmNXG1. Residues that differ in their xyloglucan interactions between EfHTG (or its homologues in other Equisetum spp.) and both PttXET and TmNXG1 (Table S3) are numbered.
(d) Two substrates modelled in EfHTG. Black carbons, XXXGXXG (a xyloglucan oligosaccharide); blue carbons, G3G4G4G4G3G4G (an MLG oligosaccharide).
(e) Cumulative enzyme–sugar interactions (see Table S3).
(f) Diagram of EfHTG and PttXET showing strong interaction (tick symbol) or weak interaction (cross) with polysaccharides and potential acceptor substrates; black and white arrowheads indicate bonds that are actually or potentially cleaved. Predicted interactions that are not present in a comparator are shown in red.
