Fig. 1.
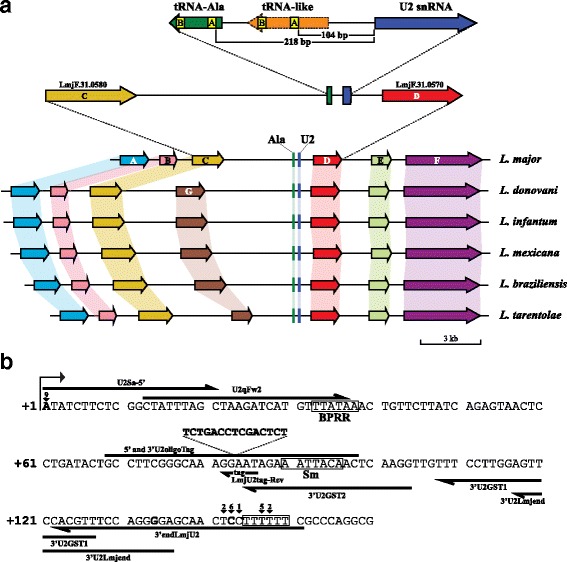
Genomic context and sequence of the U2 snRNA gene in L. major. a Synteny of U2 snRNA genes in different species of Leishmania. Orthologues genes are joined by colored lines. In L. major, gene A (light blue) corresponds to LmjF.31.0600, gene B (pink) to LmjF.31.0590, gene C (mustard) to LmjF.31.0580, gene D (red) to LmjF.31.0570, gene E (light green) to LmjF.31.0560 and gene F (purple) to LmjF.31.0550. Gene G (brown) is absent in L. major, but corresponds to LdBPK_310600.1 in L. donovani. Two enlargements of the L. major U2 snRNA gene are shown. On the top scheme are presented the U2 snRNA (blue), the tRNA-like (orange) and tRNA-Ala (green) genes. Boxes A and B within the tRNA-like and tRNA-Ala gene are shown in yellow. Distances between the U2 snRNA gene and boxes A from the tRNA-like (104 bp) and the tRNA-Ala gene (218 bp) are indicated. Figure is drawn to scale. b Sequence of the L. major U2 snRNA gene and 3'-flanking region. The A residue shown in bold type (position +1) corresponds to the TSS mapped by primer extension and 5'-RACE analyses (the arrowhead specify the number of clones found at that position). At the 3' end, the cluster of T nucleotides is shown in a box. The 3' termini mapped by RT-PCR are denoted with arrowheads, specifying the number of clones found at each position. The mature 3' terminus of the U2 snRNA (C at position +143) is shown in bold type. The missanotated 3' end of U2 at position 134 (G) is also shown in bold type. The locations of putative BPRR and Sm site are shown. The sequence of the tag, inserted between positions 83 (G) and 84 (A), is also indicated. The sequence and location of some of the oligonucleotides used in this study are shown. To facilitate the analysis of the results, the sequence is shown as the reverse-complement of the annotated sequence of chromosome 31 from the TriTrypDB database
