Fig. 4.
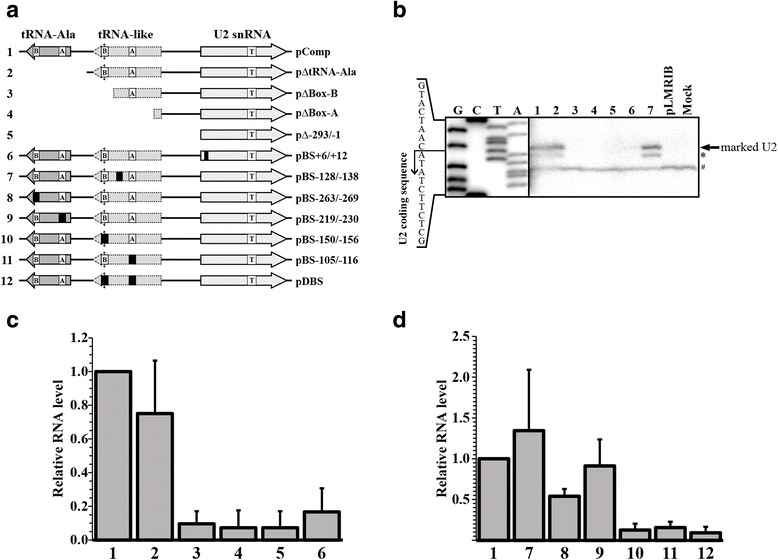
Identification of sequence elements required for the expression of the U2 snRNA. a Schematic representation of the constructs used for the transcription analysis. The location of the tag sequence (T in a box) is indicated in the U2 snRNA gene. Boxes A and B are shown in the tRNA-like and the tRNA-Ala gene. Black squares denote mutations by base substitutions. b Analysis of U2 snRNA promoter elements by primer extension assays. Constructs 1–7 were transfected into L. major cells and total RNA was obtained 24 h post-transfection. Primer extension analysis was performed with oligonucleotide LmjU2tag-Rev, which recognizes the tag sequence within the U2 exogenous transcripts. Products were separated on an 8 % polyacrylamide gel together with a DNA sequence ladder obtained with the same oligonucleotide. Lanes 1 to 7 correspond to RNA from cells transfected with vectors 1 to 7, respectively. Control experiments were carried out with RNA from mock-transfected cells (Mock), and from cells transfected with an unrelated vector (pLMRIB) [25]. c Transcriptional effect of progressive deletions of the 5'-flanking region of the U2 snRNA gene by RT-qPCR. The relative levels of the U2 snRNA were analyzed in cells transfected with constructs 1 to 6. d RT-qPCR assays to determine the effect of base substitutions in the expression of the U2 snRNA. The relative levels of U2 snRNA were analyzed in cells transfected with constructs 1 and 7–12. Error bars indicate standard deviations from three biological replicates
