Fig. 6.
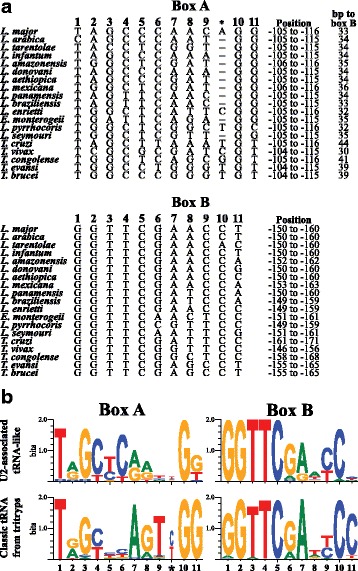
Analysis of boxes A and B from tRNA-like sequences associated to U2 snRNA genes. a Alignment of boxes A and B from the tRNA-like sequences of the following species of trypanosomatids: L. major (Friedlin), L. arabica (LEM1108), L. tarentolae (Parrot-TarII), L. infantum (JPCM5), L. amazonensis (MHOMBR/71973/M2269), L. donovani (BPK282A1), L. aethiopica (L147), L. mexicana (U1103), L. panamensis (MHOM/COL/81/L13), L. braziliensis (M2904), L. enriettii (LEM3045), E. monterogeii (LV88), L. pyrrhocoris (H10), L. seymouri (ATCC 30220), T. cruzi (CL Brener Non-Esmeraldo-like, copy of the chromosome 23), T. vivax (Y486), T. congolense (IL3000), T. evansi (STIB 805) and T. brucei (DAL972). All sequences were obtained from the TriTrypDB databases. In box A, some tRNA-like sequences contain an extra nucleotide between positions 9 and 10 (marked with an asterisk). For each box, its position in relation to the TSS of the associated U2 snRNA is indicated. The distance between boxes A and B is also shown. b Consensus sequences of boxes A and B from tRNA-like regions associated to U2 snRNA genes in trypanosomatids. The alignments shown in panel A were used to generate sequence logos with the WebLogo application (top logos). Additional sequence logos were obtained for boxes A and B from all of the 258 classic tRNAs found in TriTryps [43] (lower logos). All four sequence logos are presented by their information content
