Abstract
In this data article, the fluorescence, UV–vis absorption and FTIR spectra data of BSA-AR1/AG50 system were presented, which were used for obtaining the binding characterization (such as binding constant, binding distance, binding site, thermodynamics, and structural stability of protein) between BSA and AR1/AG50.
Keywords: Bovine serum albumin, Acid red 1, Acid green 50, Data
Specifications Table
| Subject area | Chemistry |
| More specific subject area | Analytical chemistry |
| Type of data | Table, figure |
| How data was acquired | Cary Eclipse fluorescence spectrofluorimeter (Varian, USA), UV-3600 spectrophotometer (Shimadzu, Japan), Nicolet-6700 FTIR spectrometer (Thermoscientific, USA) |
| Data format | Raw, analyzed |
| Experimental factors | The solution of BSA was prepared in phosphate-buffer (0.05 M NaH2PO4-Na2HPO4, pH=4.8, 5.5, 6.3 and 7.4) without/with salt (NaCl, 99.5% purity) or ethanol (99.7% purity) |
| Experimental features | Fluorescence, UV–vis absorption and FTIR spectra were employed to investigate binding characterization of BSA with AR1/AG50 at different conditions |
| Data source location | Nanchong, China |
| Data accessibility | Data are provided with this article |
Value of the data
-
•
The data are intuitionistic for readers to compare the binding affinity of AR1/AG50 with BSA;
-
•
The data are helpful to readers for understanding further the related parameters calculated;
-
•
The data may be of great help to study in detail the similar systems.
1. Data
The interaction data of BSA with AR1/AG50 were determined using Cary Eclipse fluorescence spectrofluorimeter (Varian, USA), UV-3600 spectrophotometer (Shimadzu, Japan) or Nicolet-6700 FTIR spectrometer; and these data were shown as fluorescence quenching spectra, the Stern–Volmer plots in the absence and presence of ethanol/NaCl, RLS spectra, UV–vis absorption spectra, UV-melting profiles, synchronous fluorescence spectra, and FTIR spectra. Corresponding parameters were calculated based on the interaction data.
In addition, to make the figures in the text become clearer, all of the figures were processed by Photoshop 8.1 software. ChemOffice 2008 was used for drawing the structures of acid red 1 and acid green 50 (Fig. 1).
Fig. 1.
The structures of acid red1 (A) and acid green 50 (B).
2. Experimental design, materials and methods
2.1. Materials
Bovine Serum Albumin (BSA, Fraction V, 98% purity, Mr= 68,000 g/mol, CAS: 9048-46-8) was purchased from Ruibio Company. AR1 (CAS: 3734-67-6) and AG50 (CAS: 3087-16-9) were obtained from J&K Scientific Ltd. (Beijing, China) and Acros Organics (New Jersey, USA), respectively. And all other chemicals were analytical reagent grade.
2.2. Methods
2.2.1. Fluorescence quenching of BSA by AR1/AG50
3.0 mL BSA solution (2.0 μM) was titrated by successive additions of AR1/AG50 solution with the concentration of 3.0×10−4 mol L−1 at different conditions (pH=4.8, 5.5, 6.3 or 7.4, T=293, 298, 304 or 310 °K, c(NaCl)=0.0, 0.04, 0.09 or 0.15 M, and/or ethanol content (%)=0%, 5% or 10%), and the final concentration of AR1/AG50 was kept at 11.54×10−6 mol L−1. The fluorescence quenching of BSA with the addition of AR1/AG50 was recorded in the range of 300–500 nm by Cary Eclipse fluorescence spectrofluorimeter (Varian, USA). The width of the excitation and emission slit was adjusted at 5 nm, and the excitation wavelength was selected at 280 nm. The temperature of samples was kept by recycle water during the whole experiment. All fluorescence titration experiments were done manually by 50 µL microsyringe.
The figures of fluorescence quenching spectra (Fig. 2) were made using Origin 7.5.
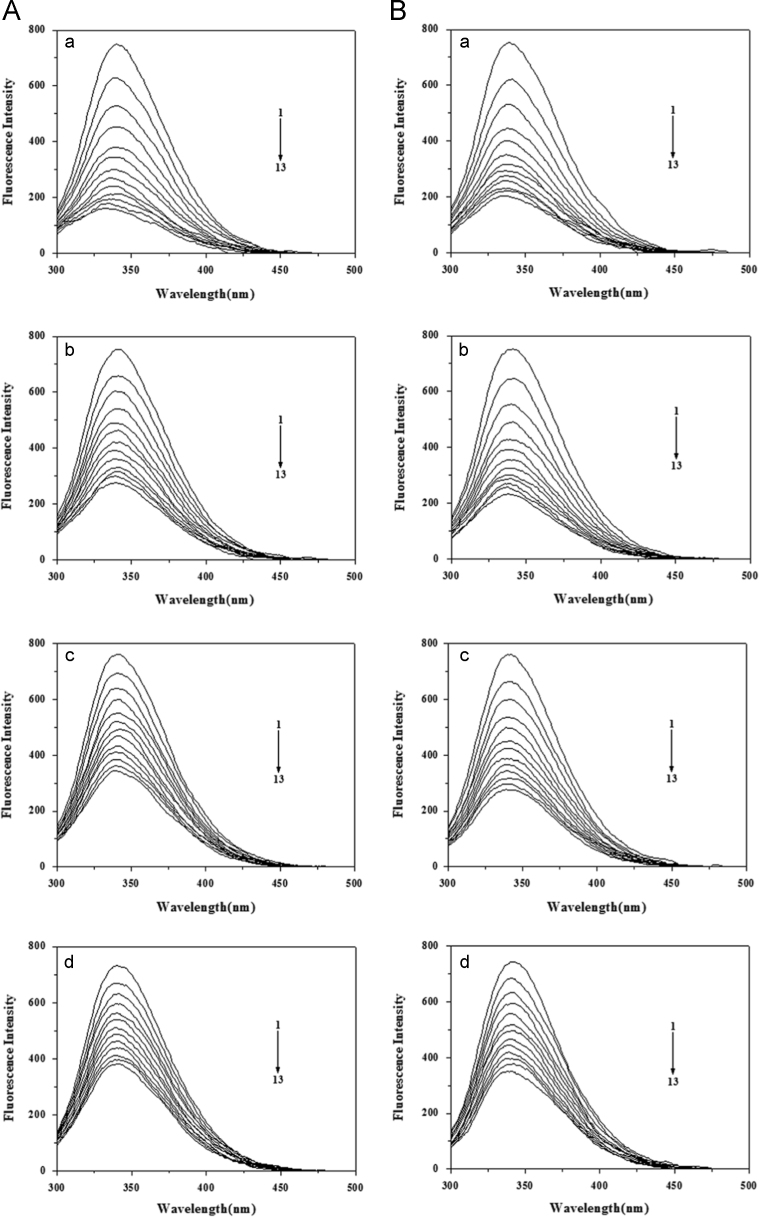
Fig. 2.
Fluorescence quenching spectra of BSA without salt by AR1 (A) or AG50 (B) at pH 4.8 (a), 5.5 (b), 6.3 (c) and 7.4 (d), respectively. λex=280 nm; c(BSA)=2.0 μM; c(AR1)=c(AG50) 1–13=0, 1.00, 1.99, 2.97, 3.95, 4.92, 5.88, 6.84, 7.79, 8.74, 9.68, 10.61, 11.54 μM; T=298 °K.
2.2.2. UV–vis absorption spectra of BSA, AR1 and AG50
Absorption spectra of AR1 (1.99 μM), AG50 (1.99 μM) and BSA (2.0 μM) in 3.0 mL phosphate-buffer was determined by UV-3600 spectrophotometer (Shimadzu, Japan) in the range of 250–350 nm, respectively; and corresponding figures (Fig. 3) were done by Origin 7.5.
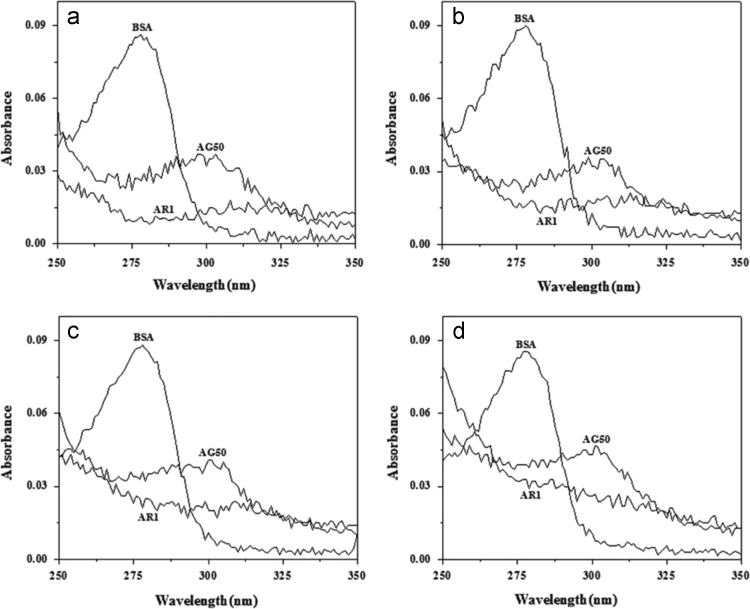
Fig. 3.
Absorption spectra of BSA, AR1 and AG50 at pH 4.8 (a), 5.5 (b), 6.3 (c) and 7.4 (d); c(BSA)=2.0 μM; c(AR1)=c(AG50)=1.99 μM; T =298 °K.
2.2.3. Spectral overlap of BSA with AR1/AG50
The fluorescence emission spectra of BSA (2.0 μM) at pH=4.8, 5.5, 6.3 or 7.4 and [NaCl]=0, 0.04, 0.09 or 0.15 M were carried out by Cary Eclipse fluorescence spectrofluorimeter, respectively. Other scanning parameters were the same as those of the fluorescence titration experiments. The UV–vis absorption spectra of AR1/AG50 (1.99 μM) were determined on UV-3600 spectrophotometer at pH=4.8, 5.5, 6.3 or 7.4 and [NaCl]=0, 0.04, 0.09 or 0.15 M, respectively. Fluorescence emission and UV–vis absorption spectra were recorded in the range of 250–500 nm.
The figures of spectral overlap between BSA and AR1/AG50 (Fig. 4) were done by Origin 7.5.
Fig. 4.
Spectral overlap of UV–vis absorption spectrum of AR1 or AG50 with the fluorescence emission spectrum of BSA at pH 4.8, 5.5, 6.3 or 7.4; c(BSA)=2.0 μM, c(AR1)=c(AG50)=1.99 μM, T=298 °K.
2.2.4. The S–V plots of BSA-AR1/AG50 system
The measured fluorescence quenching data of BSA by AR1/AG50 at different conditions were corrected [1] and fitted by Origin 7.5 based on Eq. (1) (Fig. 5), and corresponding values were listed in Table 1.
| (1) |
Fig. 5.
The S–V plots for the quenching of BSA by AR1 (A and C) or AG50 (B and D) at pH=4.8 (A and B) and 7.4 (C and D); λex=280 nm.
Table 1.
The parameters of S–V plot for the BSA-AR1/AG50system at different conditions.
| pH | NaCl (M) | T (K) | BSA-AR1 |
BSA-AG50 |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| KSV (×10−4 M−1) | Kq (×10−12 M−1 s−1) | R | SD | KSV (×10−4 M−1) | Kq (×10−12 M−1 s−1) | R | SD | |||
| 4.8 | 0 | 293 | 32.88 | 32.88 | 0.9973 | 0.0887 | 18.58 | 18.58 | 0.9969 | 0.0528 |
| 298 | 27.17 | 27.17 | 0.9978 | 0.0655 | 16.68 | 16.68 | 0.9979 | 0.0393 | ||
| 304 | 23.75 | 23.75 | 0.9981 | 0.0532 | 14.16 | 14.16 | 0.9997 | 0.0130 | ||
| 310 | 19.43 | 19.43 | 0.9990 | 0.0322 | 11.56 | 11.56 | 0.9982 | 0.0249 | ||
| 0.04 | 293 | 17.11 | 17.11 | 0.9988 | 0.0305 | 13.07 | 13.07 | 0.9995 | 0.0154 | |
| 298 | 14.37 | 14.37 | 0.9992 | 0.0206 | 10.41 | 10.41 | 0.9993 | 0.0139 | ||
| 304 | 13.55 | 13.55 | 0.9988 | 0.0241 | 8.79 | 8.79 | 0.9993 | 0.0119 | ||
| 310 | 11.74 | 11.74 | 0.9986 | 0.0229 | 7.00 | 7.00 | 0.9996 | 0.0070 | ||
| 0.09 | 293 | 12.58 | 12.58 | 0.9990 | 0.0205 | 8.67 | 8.67 | 0.9999 | 0.0036 | |
| 298 | 10.52 | 10.52 | 0.9985 | 0.0209 | 7.97 | 7.97 | 0.9987 | 0.0147 | ||
| 304 | 9.00 | 9.00 | 0.9975 | 0.0232 | 6.72 | 6.72 | 0.9998 | 0.0045 | ||
| 310 | 7.19 | 7.19 | 0.9991 | 0.0112 | 5.85 | 5.85 | 0.9989 | 0.0098 | ||
| 0.15 | 293 | 9.74 | 9.74 | 0.9990 | 0.0157 | 6.52 | 6.52 | 0.9993 | 0.0089 | |
| 298 | 8.02 | 8.02 | 0.9980 | 0.0185 | 5.66 | 5.66 | 0.9996 | 0.0062 | ||
| 304 | 6.76 | 6.76 | 0.9990 | 0.0108 | 4.96 | 4.96 | 0.9996 | 0.0048 | ||
| 310 | 5.97 | 5.97 | 0.9977 | 0.0148 | 4.41 | 4.41 | 0.9992 | 0.0063 | ||
| 7.4 | 0 | 293 | 7.76 | 7.76 | 0.9995 | 0.0086 | 6.46 | 6.46 | 0.9992 | 0.0097 |
| 298 | 6.13 | 6.13 | 0.9988 | 0.0111 | 5.81 | 5.81 | 0.9992 | 0.0084 | ||
| 304 | 5.22 | 5.22 | 0.9993 | 0.0073 | 4.79 | 4.79 | 0.9985 | 0.0095 | ||
| 310 | 4.59 | 4.59 | 0.9987 | 0.0084 | 3.77 | 3.77 | 0.9990 | 0.0062 | ||
| 0.04 | 293 | 5.01 | 5.01 | 0.9972 | 0.0136 | 4.20 | 4.20 | 0.9995 | 0.0051 | |
| 298 | 3.76 | 3.76 | 0.9955 | 0.0130 | 3.75 | 3.75 | 0.9992 | 0.0055 | ||
| 304 | 3.59 | 3.59 | 0.9990 | 0.0058 | 3.58 | 3.58 | 0.9989 | 0.0061 | ||
| 310 | 3.25 | 3.25 | 0.9995 | 0.0038 | 3.22 | 3.22 | 0.9992 | 0.0046 | ||
| 0.09 | 293 | 3.84 | 3.84 | 0.9976 | 0.0096 | 3.81 | 3.81 | 0.9984 | 0.0077 | |
| 298 | 3.59 | 3.59 | 0.9969 | 0.0103 | 3.58 | 3.58 | 0.9979 | 0.0084 | ||
| 304 | 3.14 | 3.14 | 0.9982 | 0.0068 | 3.07 | 3.07 | 0.9981 | 0.0068 | ||
| 310 | 2.91 | 2.91 | 0.9985 | 0.0058 | 2.87 | 2.87 | 0.9971 | 0.0080 | ||
| 0.15 | 293 | 3.10 | 3.10 | 0.9976 | 0.0078 | 2.39 | 2.39 | 0.9984 | 0.0050 | |
| 298 | 2.90 | 2.90 | 0.9981 | 0.0066 | 2.25 | 2.25 | 0.9993 | 0.0030 | ||
| 304 | 2.60 | 2.60 | 0.9989 | 0.0044 | 2.13 | 2.13 | 0.9994 | 0.0027 | ||
| 310 | 2.29 | 2.29 | 0.9986 | 0.0044 | 1.90 | 1.90 | 0.9976 | 0.0048 | ||
R and SD are the correlation coefficient and the standard deviation for the S–V plots, respectively.
2.2.5. RLS spectra of BSA-AR1/AG50 system
RLS spectra of BSA (2.0 μM) with the addition of AR1/AG50 (0-11.54 μM) at Δλ=0 nm were determined in the range of 250–700 nm by Cary Eclipse fluorescence spectrofluorimeter at pH 4.8 and 7.4, respectively; and corresponding figures of RLS spectra (Fig. 6) were done by Origin 7.5. Other scanning parameters were the same as those of the fluorescence titration experiments.
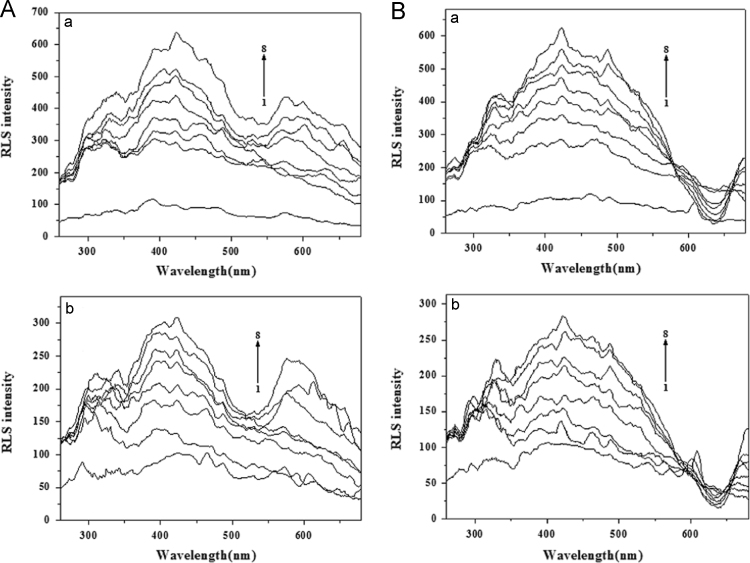
Fig. 6.
RLS spectra of the BSA-AR1 (A) and BSA-AG50 (B) systems at pH 4.8 (a) and 7.4 (b). Curve 1: only AR1 or AG50 (1.99 μM); curve 2: only BSA (2.0 μM); c(AR1)=c(AG50) 3–8=1.99, 3.95, 5.88, 7.79, 9.68, 11.54 μM.
2.2.6. UV–vis absorption spectra of BSA-AR1/AG50 system
UV–vis absorption spectra (in the range of 250–400 nm) of BSA (2.0 μM) in the presence of AR1/AG50 (0–11.54 μM) at pH 4.8, 5.5, 6.3 or 7.4 were determined on UV-3600 spectrophotometer, and their figures (Fig. 7) were made by Origin 7.5.
Fig. 7.
UV–vis absorption spectra of BSA in the presence of AR1 (A) or AG50 (B) at pH 4.8 (a), 5.5 (b), 6.3 (c) and 7.4 (b); c(BSA)=2.0 μM; c(AR1)=c(AG50) 1–13=0, 1.00, 1.99, 2.97, 3.95, 4.92, 5.88, 6.84, 7.79, 8.74, 9.68, 10.61, 11.54 μM; T=298 °K.
2.2.7. The effect of ethanol on the quenching plots of BSA-AR1/AG50 system
The measured fluorescence quenching data of BSA by AR1/AG50 without or with ethanol content (5% or 10%) at pH=4.8, 5.5, 6.3 and 7.4 were corrected [1] and fitted by Origin 7.5 based on Eq. (1) (Fig. 8).
Fig. 8.
Effects of increasing ethanol content on the quenching plots of BSA by AR1 (A) or AG50 (B) at different pH values, T=298 °K, λex=280 nm.
2.2.8. The effect of NaCl on the absorption spectra of BSA-AR1/AG50 system
The absorption spectra of AR1 (42 μM) or AG50 (8 μM) without or with NaCl concentration (0.04, 0.09 or 0.15 M) in the presence of BSA (0–60 μM) were recorded in the range of 300–600 nm by an UV-3600 spectrophotometer at pH 4.8 and 7.4, respectively; and corresponding absorption spectra were fitted using Origin 7.5 based on Eq. (2) (Fig. 9).
| (2) |
Fig. 9.
Absorption spectra of AR1 or AG50 with increasing the concentrations of BSA and Benesi–Hildebrand plots for the BSA-AR1 (A and B) or BSA-AG50 (C and D) complex in the presence of different salt concentrations at pH 4.8 and 7.4; c(AR1)=42 μM, c(AG50)=8 μM, c(BSA) 1–13=0, 5, 10, 15, 20, 25, 30, 35, 40, 45, 50, 55, 60 μM, T=298 °K.
2.2.9. The effect of NaCl on the quenching plots of BSA-AR1/AG50 system
The measured fluorescence quenching data of BSA by AR1/AG50 without or with NaCl (0.04, 0.09 or 0.15 M) at T=293, 298, 304 or 310 °K and pH=4.8 or 7.4 were corrected [1] and fitted by Origin 7.5 based on Eq. (1) (Fig. 10).
Fig. 10.
Effect of increasing NaCl concentrations on the quenching plots of BSA by AR1 (A and C) or AG50 (B and D) at pH 4.8 (A and B) or 7.4 (C and D), λex=280 nm.
2.2.10. The kinetics of BSA-AR1/AG50 system
The absorption spectra of BSA-AR1/AG50 system (T=293, 298, 304 and 310 °K, pH=4.8 and 7.4) (Fig. 11A) were measured at 278 nm by an UV-3600 spectrophotometer at different time intervals. And semilogarithmic plots of ln[(Amax−At)/A0] vs. incubation time (t) for the BSA-AR1/AG50 system (Fig. 11B) were done by Origin 7.5.
Fig. 11.
Plots (A) of BSA-AR1 (a and b) or BSA-AG50 (c and d) absorption vs. incubation time (t) and plots (B) of ln[(Amax−At)/A0] vs. incubation time (t) for the BSA-AR1 (a and b) or BSA-AG50 (c and d) system at different temperatures or pH. c(BSA)=2.0 μM, c(AR1)=c(AG50)=49.59 μM.
2.2.11. The UV-melting profiles of BSA-AR1/AG50 system
Free BSA (5.0 μM) or BSA-AR1/AG50 (39.74 μM) complex without or with NaCl (0.15 M) were monitored at 278 nm by UV-3600 spectrophotometer with increasing temperature (from 25 to 100 °C with a rate of 1 °C/min). And their UV-melting profiles (Fig. 12) were fitted by MTLAB 2010.
Fig. 12.
UV-melting profiles (absorbance change vs. temperature at 278 nm) of BSA and BSA-AR1 (A) or BSA-AG50 (B) system; curve a, b or c represents pattern for free BSA, BSA-AR1/AG50 or BSA-AR1/AG50 with NaCl (0.15 M). c(BSA)=5.0 μM, c(AR1)=c(AG50)=39.74 μM, pH=4.8.
2.2.12. The synchronous fluorescence spectra
Synchronous fluorescence spectra of BSA (2.0 μM) with the increasing AR1/AG50 concentration (0–75.00 μM) at Δλ=15 and 60 nm were recorded using Cary Eclipse fluorescence spectrofluorimeter in the range of 250–500 nm. Other scanning parameters were the same as those of the fluorescence titration experiments. Besides, corresponding figures (Fig. 13) was made by Origin 7.5.
Fig. 13.
Synchronous fluorescence spectra of BSA-AR1 (A and C) or BSA-AG50 (B and D) system at Δλ=15 and 60 nm; c(BSA)=2.0 μM, c(AR1)=c(AG50) 1–7=0, 1.99, 3.95, 5.88, 7.79, 9.68, 11.54 μM; T=298 °K.
2.2.13. FTIR spectra
FTIR spectra of free BSA (0.2 mM), BSA-AR1 and BSA-AG50 complexes (the molar ratio of BSA to AR1 or AG50 is maintained at 1:1) were recorded on Nicolet-6700 FTIR spectrometer via the attenuated total reflection (ATR) at a resolution of 4 cm−1 and 64 scans in the range of 400–4000 cm−1 at room temperature. The corresponding absorbance contributions of buffer and free AR1/AG50 solutions were recorded and digitally subtracted with the same instrumental parameters, and their FTIR spectra (Fig. 14) was done by OMNIC.
Fig. 14.
FTIR spectra in the 1800–900 cm−1 region for free BSA (0.2 mM), BSA-AR1 and BSA-AG50 complexes (the molar ratio of BSA to AR1 or AG50 is maintained at 1:1), and their corresponding difference spectra were indicated in the figure. The contribution of AR1 or AG50 is subtracted from the different spectra in this region.
2.2.14. The parameters of S–V plot
The parameters of fluorescence quenching for the BSA-AR1/AG50 system at different conditions were calculated using the S–V equation [1].
2.2.15. Effect of pH, NaCl and ethanol on the binding parameters of BSA-AR1/AG50 system
The binding parameters of the two systems (Tables 2–4) were calculated using double logarithm regression curves, Debye–Hückel limiting law and Van’t Hoff equation based on the data of fluorescence quenching at different conditions, respectively [2], [3].
Table 2.
Effect of pH on the parameters calculated by Debye–Hückel limiting law for the BSA-AR1/AG50 system.
| Ligind | T (K) | I1/2 | pH 4.8 |
I1/2 | pH 7.4 |
ΔZP=ZP,pH4.8-ZP,pH7.4 | ||||||||||
| log(Kexp.1) | log(Kexp.2) | log(Kexp.3) | log(K) | Keq (×10−3 M−1) | ZP | log(Kexp.1) | log(Kexp.2) | log(Kexp.3) | log(K) | Keq (×10−3 M−1) | ZP | |||||
| AR1 | 293 | 0.22 | 6.21401 | 6.00249 | 6.08425 | 6.10025 | 4378.9 | 1.18 | 0.26 | 4.45338 | 4.25311 | 4.32311 | 4.34320 | 33.6 | 0.35 | 0.83 |
| 0.30 | 6.01412 | 5.79428 | 5.90369 | 5.90403 | 0.33 | 4.38801 | 4.18799 | 4.25857 | 4.27819 | |||||||
| 0.37 | 5.83139 | 5.61051 | 5.73388 | 5.72526 | 0.40 | 4.35407 | 4.15423 | 4.22478 | 4.24436 | |||||||
| 0.45 | 5.64213 | 5.42627 | 5.53885 | 5.53575 | 0.47 | 4.29632 | 4.09638 | 4.16689 | 4.18653 | |||||||
| 298 | 0.22 | 6.15317 | 5.93821 | 6.04122 | 6.04420 | 4210.2 | 1.30 | 0.26 | 4.38491 | 4.18479 | 4.25536 | 4.27502 | 30.9 | 0.41 | 0.89 | |
| 0.30 | 5.95846 | 5.73241 | 5.84623 | 5.84570 | 0.33 | 4.32073 | 4.12087 | 4.19098 | 4.21086 | |||||||
| 0.37 | 5.72893 | 5.52828 | 5.62019 | 5.62580 | 0.40 | 4.29111 | 4.09109 | 4.16146 | 4.18122 | |||||||
| 0.45 | 5.55324 | 5.33887 | 5.45339 | 5.44850 | 0.47 | 4.20272 | 4.00248 | 4.07338 | 4.09286 | |||||||
| 304 | 0.22 | 6.09431 | 5.87959 | 5.97375 | 5.98255 | 4023.8 | 1.42 | 0.26 | 4.30889 | 4.09871 | 4.18949 | 4.19903 | 25.6 | 0.40 | 1.02 | |
| 0.30 | 5.89338 | 5.67842 | 5.78608 | 5.78596 | 0.33 | 4.25238 | 4.05222 | 4.12254 | 4.14238 | |||||||
| 0.37 | 5.64212 | 5.42808 | 5.54008 | 5.53676 | 0.40 | 4.22402 | 4.02478 | 4.09677 | 4.11519 | |||||||
| 0.45 | 5.46171 | 5.24669 | 5.35129 | 5.35323 | 0.47 | 4.13521 | 3.93449 | 4.00350 | 4.02440 | |||||||
| 310 | 0.22 | 6.02188 | 5.80111 | 5.92981 | 5.91760 | 3578.4 | 1.50 | 0.26 | 4.24029 | 4.04021 | 4.10992 | 4.13014 | 22.8 | 0.44 | 1.06 | |
| 0.30 | 5.83412 | 5.61538 | 5.73792 | 5.72914 | 0.33 | 4.20342 | 4.00328 | 4.07386 | 4.09352 | |||||||
| 0.37 | 5.55041 | 5.36749 | 5.41161 | 5.44317 | 0.40 | 4.13913 | 3.93847 | 4.00851 | 4.02870 | |||||||
| 0.45 | 5.38409 | 5.16861 | 5.30047 | 5.28439 | 0.47 | 4.06603 | 3.86537 | 3.93603 | 3.95581 | |||||||
| AG50 | 293 | 0.22 | 5.17015 | 4.97229 | 5.04515 | 5.06253 | 162.1 | 0.66 | 0.26 | 4.32163 | 4.12127 | 4.19274 | 4.21188 | 20.2 | 0.34 | 0.32 |
| 0.30 | 5.10342 | 4.91228 | 4.99252 | 5.00274 | 0.33 | 4.29691 | 4.09679 | 4.16763 | 4.18711 | |||||||
| 0.37 | 5.06033 | 4.85987 | 4.93082 | 4.95034 | 0.40 | 4.27288 | 4.07312 | 4.14321 | 4.16307 | |||||||
| 0.45 | 5.01017 | 4.81032 | 4.89901 | 4.90650 | 0.47 | 4.24649 | 4.04681 | 4.11746 | 4.13692 | |||||||
| 298 | 0.22 | 5.13221 | 4.93139 | 5.01515 | 5.02625 | 154.6 | 0.74 | 0.26 | 4.27812 | 4.07788 | 4.14908 | 4.16836 | 18.8 | 0.42 | 0.32 | |
| 0.30 | 5.06133 | 4.86007 | 4.95539 | 4.95893 | 0.33 | 4.23901 | 4.03919 | 4.10982 | 4.12934 | |||||||
| 0.37 | 5.02049 | 4.82311 | 4.89861 | 4.91407 | 0.40 | 4.20907 | 4.00913 | 4.07949 | 4.09923 | |||||||
| 0.45 | 4.96028 | 4.75232 | 4.84058 | 4.85106 | 0.47 | 4.18849 | 3.98841 | 4.05953 | 4.07881 | |||||||
| 304 | 0.22 | 5.09308 | 4.89562 | 4.96882 | 4.98584 | 139.0 | 0.74 | 0.26 | 4.23508 | 4.02812 | 4.06773 | 4.11031 | 16.2 | 0.41 | 0.33 | |
| 0.30 | 5.02351 | 4.82609 | 4.90182 | 4.91714 | 0.33 | 4.17893 | 3.97827 | 4.05037 | 4.06919 | |||||||
| 0.37 | 4.98413 | 4.78257 | 4.85455 | 4.87375 | 0.40 | 4.12771 | 3.93549 | 4.02959 | 4.03093 | |||||||
| 0.45 | 4.92152 | 4.72447 | 4.79961 | 4.81520 | 0.47 | 4.12221 | 3.93048 | 4.02951 | 4.02740 | |||||||
| 310 | 0.22 | 5.05412 | 4.85208 | 4.92463 | 4.94361 | 128.9 | 0.79 | 0.26 | 4.19188 | 3.99182 | 4.06278 | 4.08216 | 16.9 | 0.62 | 0.17 | |
| 0.30 | 4.98051 | 4.78229 | 4.86229 | 4.87503 | 0.33 | 4.13839 | 3.93851 | 4.00956 | 4.02882 | |||||||
| 0.37 | 4.94652 | 4.74648 | 4.82031 | 4.83777 | 0.40 | 4.08361 | 3.88339 | 3.95458 | 3.97386 | |||||||
| 0.45 | 4.87117 | 4.67391 | 4.74542 | 4.76350 | 0.47 | 4.07203 | 3.87187 | 3.94297 | 3.96229 | |||||||
| Ligind | T (K) | I1/2 | pH 4.8 |
I1/2 | pH 7.4 |
||||||||
| (kJ mol−1) | (kJ mol−1) | (kJ mol−1) | ΔG° (kJ mol−1) | (kJ mol−1) | (kJ mol−1) | (kJ mol−1) | (kJ mol−1) | ΔG° (kJ mol−1) | (kJ mol−1) | ||||
| AR1 | 293 | 0.22 | −34.9 | −33.7 | −34.1 | −34.2 | −37.3 | 0.26 | −25.0 | −23.7 | −24.2 | −24.3 | −25.4 |
| 0.30 | −33.7 | −32.5 | −33.1 | −33.1 | 0.33 | −24.6 | −23.5 | −23.9 | −24.0 | ||||
| 0.37 | −32.7 | −31.4 | −32.1 | −32.1 | 0.40 | −24.4 | −23.3 | −23.7 | −23.8 | ||||
| 0.45 | −31.6 | −30.4 | −31.0 | −31.0 | 0.47 | −23.8 | −23.3 | −23.2 | −23.3 | ||||
| 298 | 0.22 | −35.1 | −33.9 | −34.4 | −34.5 | −37.8 | 0.26 | −25.0 | −23.9 | −24.3 | −24.4 | −25.6 | |
| 0.30 | −34.0 | −32.7 | −33.4 | −33.4 | 0.33 | −24.7 | −23.5 | −23.9 | −24.0 | ||||
| 0.37 | −32.7 | −31.5 | −32.1 | −32.1 | 0.40 | −24.5 | −23.3 | −23.7 | −23.8 | ||||
| 0.45 | −31.7 | −30.5 | −31.1 | −31.1 | 0.47 | −24.0 | −23.3 | −23.2 | −23.4 | ||||
| 304 | 0.22 | −35.4 | −34.2 | −34.8 | −34.8 | −38.4 | 0.26 | −25.1 | −23.9 | −24.3 | −24.4 | −25.7 | |
| 0.30 | −34.3 | −33.0 | −33.7 | −33.7 | 0.33 | −24.8 | −23.6 | −24.0 | −24.2 | ||||
| 0.37 | −32.8 | −31.7 | −32.1 | −32.2 | 0.40 | −24.5 | −23.4 | −23.8 | −23.9 | ||||
| 0.45 | −31.8 | −30.6 | −31.3 | −31.2 | 0.47 | −24.0 | −23.3 | −23.2 | −23.4 | ||||
| 310 | 0.22 | −35.8 | −34.4 | −35.2 | −35.1 | −39.0 | 0.26 | −25.2 | −23.9 | −24.4 | −24.5 | −25.9 | |
| 0.30 | −34.6 | −33.3 | −34.0 | −34.0 | 0.33 | −24.9 | −23.7 | −24.1 | −24.3 | ||||
| 0.37 | −32.9 | −31.8 | −32.2 | −32.3 | 0.40 | −24.6 | −23.4 | −23.8 | −23.9 | ||||
| 0.45 | −31.9 | −30.6 | −31.4 | −31.3 | 0.47 | −24.0 | −23.3 | −23.2 | −23.4 | ||||
| AG50 | 293 | 0.22 | −29.0 | −27.9 | −28.3 | −28.4 | −29.2 | 0.22 | −24.2 | −23.1 | −23.5 | −23.6 | −24.1 |
| 0.30 | −28.6 | −27.5 | −28.0 | −28.0 | 0.30 | −24.1 | −23.0 | −23.3 | −23.5 | ||||
| 0.37 | −28.4 | −27.3 | −27.7 | −27.8 | 0.37 | −23.9 | −22.8 | −23.2 | −23.3 | ||||
| 0.45 | −28.1 | −27.0 | −27.5 | −27.5 | 0.45 | −23.8 | −22.7 | −23.1 | −23.2 | ||||
| 298 | 0.22 | −29.3 | −28.2 | −28.6 | −28.7 | −29.6 | 0.22 | −24.4 | −23.3 | −23.6 | −23.8 | −24.4 | |
| 0.30 | −28.9 | −27.8 | −28.3 | −28.3 | 0.30 | −24.2 | −23.1 | −23.5 | −23.6 | ||||
| 0.37 | −28.7 | −27.5 | −27.9 | −28.0 | 0.37 | −24.0 | −22.9 | −23.3 | −23.4 | ||||
| 0.45 | −28.3 | −27.2 | −27.7 | −27.7 | 0.45 | −23.9 | −22.8 | −23.2 | −23.3 | ||||
| 304 | 0.22 | −29.6 | −28.5 | −28.9 | −29.0 | −30.0 | 0.22 | −24.6 | −23.5 | −23.8 | −24.0 | −24.7 | |
| 0.30 | −29.2 | −28.1 | −28.5 | −28.6 | 0.30 | −24.4 | −23.2 | −23.6 | −23.7 | ||||
| 0.37 | −29.0 | −27.8 | −28.3 | −28.4 | 0.37 | −24.1 | −22.9 | −23.4 | −23.5 | ||||
| 0.45 | −28.6 | −27.5 | −27.9 | −28.0 | 0.45 | −24.0 | −22.8 | −23.2 | −23.3 | ||||
| 310 | 0.22 | −30.0 | −28.8 | −29.2 | −29.3 | −30.3 | 0.22 | −24.9 | −23.7 | −24.0 | −24.2 | −25.0 | |
| 0.30 | −29.6 | −28.4 | −28.8 | −28.9 | 0.30 | −24.5 | −23.3 | −23.8 | −23.9 | ||||
| 0.37 | −29.3 | −28.2 | −28.6 | −28.7 | 0.37 | −24.2 | −23.0 | −23.5 | −23.6 | ||||
| 0.45 | −28.9 | −27.7 | −28.2 | −28.3 | 0.45 | −24.1 | −23.0 | −23.5 | −23.5 | ||||
Kexp1, Kexp2 and Kexp3 denote the binding constants of BSA-AR1/AG50 system in triplicate, respectively; corresponding change of free energy (ΔG°) is , and ; log (K) and ΔG° express the average values of log (Kexp1), log (Kexp2) and log (Kexp3), and , and , respectively.
Table 4.
Effects of ethanol or pH on the binding parameters of BSA-AR1/AG50 system.
| Systems | ethanol (v/v) | parameters | pH |
|||
|---|---|---|---|---|---|---|
| 4.8 | 5.5 | 6.3 | 7.4 | |||
| BSA-AR1 | 0 | K (×10−3 M−1) (×10−3 L mol−1) | 1107.06 | 169.29 | 59.65 | 18.84 |
| n | 1.13 | 1.03 | 0.97 | 0.90 | ||
| R | 0.9995 | 0.9996 | 0.9987 | 0.9958 | ||
| 5 | K (×10−3 M−1) (×10−3 L mol−1) | 553.22 | 42.99 | 36.33 | 14.16 | |
| n | 1.13 | 0.96 | 0.98 | 0.89 | ||
| R | 0.9997 | 0.9984 | 0.9998 | 0.9964 | ||
| 10 | K (×10−3 M−1) (×10−3 L mol−1) | 169.53 | 27.72 | 25.20 | 10.88 | |
| n | 1.07 | 0.97 | 0.99 | 0.90 | ||
| R | 0.9997 | 0.9978 | 0.9954 | 0.9931 | ||
| BSA-AG50 | 0 | K (×10−3 M−1) (×10−3 L mol−1) | 106.23 | 84.31 | 51.00 | 14.74 |
| n | 0.96 | 0.95 | 0.94 | 0.88 | ||
| R | 0.9990 | 0.9993 | 0.9990 | 0.9990 | ||
| 5 | K (×10−3 M−1) (×10−3 L mol−1) | 57.08 | 36.20 | 26.76 | 7.25 | |
| n | 0.98 | 0.98 | 0.96 | 0.89 | ||
| R | 0.9991 | 0.9966 | 0.9945 | 0.9997 | ||
| 10 | K (×10−3 M−1) (×10−3 L mol−1) | 32.95 | 20.11 | 16.61 | 5.59 | |
| n | 1.01 | 0.99 | 1.01 | 0.90 | ||
| R | 0.9995 | 0.9952 | 0.9981 | 0.9954 | ||
2.2.16. The binding distances for the BSA-AR1/AG50system
According to Fig. 4, the binding distances of BSA-AR1/AG50 system at different conditions (Table 5) were calculated by Föster׳s non-radiative energy transfer theory [4].
Table 5.
The binding distances for the BSA-AR1/AG50 system at different pH and salt concentrations.
| Systems | pH |
r (nm) |
|||
|---|---|---|---|---|---|
| 0.0 M NaCl | 0.04 M NaCl | 0.09 M NaCl | 0.15 M NaCl | ||
| BSA-AR1 | 4.8 | 2.62 | 2.84 | 3.05 | 3.18 |
| 5.5 | 2.99 | 3.07 | 3.42 | 3.55 | |
| 6.3 | 3.19 | 3.66 | 3.72 | 3.85 | |
| 7.4 | 3.38 | 3.72 | 3.75 | 3.91 | |
| BSA-AG50 | 4.8 | 2.91 | 3.17 | 3.42 | 3.55 |
| 5.5 | 3.01 | 3.23 | 3.46 | 3.57 | |
| 6.3 | 3.21 | 3.68 | 3.76 | 3.88 | |
| 7.4 | 3.45 | 3.74 | 3.80 | 3.92 | |
2.2.17. The values of KB–H and ΔGB–H for the BSA-AR1/AG50 system
According to Fig. 9, the values of KB–H and ΔGB–H for the BSA-AR1/AG50 complex (Table 6) were calculated using Eq. (2).
Table 6.
The values of KB–H and ΔGB–H for the BSA-AR1/AG50 complex at different conditions.
| Systems | pH | Parameters | NaCl (M) |
|||
|---|---|---|---|---|---|---|
| 0 | 0.04 | 0.09 | 0.15 | |||
| BSA-AR1 | 4.8 | KB–H (×10−3 L mol−1) | 158.62 | 139.37 | 77.02 | 65.45 |
| R | 0.9866 | 0.9962 | 0.9963 | 0.9975 | ||
| ΔGB–H (kJ mol−1) | −29.67 | −29.35 | −27.88 | −10.35 | ||
| 7.4 | KB–H (×10−3 L mol−1) | 45.00 | 38.05 | 35.20 | 23.28 | |
| R | 0.9999 | 0.9993 | 0.9983 | 0.9994 | ||
| ΔGB–H (kJ mol−1) | −9.43 | −9.02 | −8.82 | −7.80 | ||
| BSA-AG50 | 4.8 | KB–H (×10−3 L mol−1) | 148.48 | 82.99 | 47.08 | 41.65 |
| R | 0.9871 | 0.9998 | 0.9978 | 0.9989 | ||
| ΔGB–H (kJ mol−1) | −12.39 | −10.95 | −9.54 | −9.24 | ||
| 7.4 | KB–H (×10−3 L mol−1) | 36.12 | 15.24 | 10.54 | 3.94 | |
| R | 0.9870 | 0.9980 | 0.9954 | 0.9974 | ||
| ΔGB–H (kJ mol−1) | −8.89 | −6.75 | −5.84 | −3.40 | ||
2.2.18. Kinetics study of the BSA-AR1/AG50 system
The binding rate constants (k) for the BSA-AR1/AG50 system were calculated based on the data from Fig. 11 (Table 7).
Table 7.
The binding rate constants k and corresponding statistical parameters for the BSA-AR1/AG50 system at different conditions.
| Systems | NaCl (M) | T (K) | pH 4.8 |
pH 7.4 |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| kexp.1 (×103 min−1) | kexp.2 (×103 min−1) | kexp.3 (×103 min−1) | k (×103 min−1) | R | SD | kexp.1 (×103 min−1) | kexp.2 (×103 min−1) | kexp.3 (×103 min−1) | k (×103 min−1) | R | SD | |||
| BSA-AR1 | 0 | 293 | 150.4 | 157.9 | 154.3 | 154.2 | 0.9708 | 0.2423 | 99.6 | 106.8 | 102.6 | 103.0 | 0.9858 | 0.1416 |
| 298 | 223.9 | 227.4 | 229.4 | 226.9 | 0.9699 | 0.2307 | 194.2 | 198.4 | 195.1 | 195.9 | 0.9658 | 0.2509 | ||
| 304 | 410.7 | 418.6 | 418.4 | 415.9 | 0.9804 | 0.2658 | 218.4 | 225.8 | 221.2 | 221.8 | 0.9643 | 0.2116 | ||
| 310 | 506.4 | 513.2 | 508.9 | 509.5 | 0.9733 | 0.3134 | 374.2 | 379.9 | 377.5 | 377.2 | 0.9746 | 0.2262 | ||
| 0.15 | 298 | 179.9 | 188.2 | 189.5 | 185.9 | 0.9651 | 0.1897 | 168.9 | 173.5 | 175.4 | 172.6 | 0.9666 | 0.1919 | |
| BSA-AG50 | 0 | 293 | 117.9 | 125.1 | 124.2 | 122.4 | 0.9691 | 0.2429 | 96.7 | 105.8 | 98.7 | 100.4 | 0.9848 | 0.1613 |
| 298 | 178.2 | 184.6 | 180.2 | 181.0 | 0.9939 | 0.0869 | 160.8 | 164.1 | 168.3 | 164.4 | 0.9745 | 0.2186 | ||
| 304 | 305.1 | 311.2 | 305.9 | 307.4 | 0.9704 | 0.2435 | 205.1 | 210.6 | 205.3 | 207.0 | 0.9780 | 0.1914 | ||
| 310 | 414.8 | 425.3 | 414.2 | 418.1 | 0.9840 | 0.1973 | 261.4 | 269.8 | 265.9 | 265.7 | 0.9782 | 0.1632 | ||
| 0.15 | 298 | 158.1 | 166.3 | 168.8 | 164.4 | 0.9866 | 0.1817 | 124.3 | 134.6 | 126.9 | 128. 6 | 0.9598 | 0.2607 | |
R and SD are the correlation coefficient and the standard deviation for k values, respectively; kexp.1,kexp.2 and kexp.3 denote the binding rate constants of the BSA-AR1/AG50 system in triplicate, respectively; k is the average values of kexp.1,kexp.2 and kexp.3.
Table 3.
The binding constants K, binding sites number n and thermodynamic parameters for the BSA-AR1/AG50 system at different conditions.
| Systems | NaCl (M) | T (K) | pH4.8 |
pH 7.4 |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| K (×10−3 M−1) | n | Ra | ΔG° (kJ mol−1) | ΔH° (kJ mol−1) | ΔS° (J mol−1 K−1) | Rb | K (×10−3 M−1) | n | Ra | ΔG° (kJ mol−1) | ΔH° (kJ mol−1) | ΔS° (J mol−1 K−1) | Rb | |||
| BSA-AR1 | 0 | 293 | 1259.65 | 1.13 | 0.9981 | −34.22 | −18.59 | 53.34 | 0.9998 | 22.04 | 0.89 | 0.9990 | −24.35 | −21.80 | 8.71 | 0.9998 |
| 298 | 1107.06 | 1.13 | 0.9995 | −34.48 | 18.84 | 0.90 | 0.9958 | −24.40 | ||||||||
| 304 | 960.62 | 1.13 | 0.9997 | −34.80 | 15.81 | 0.89 | 0.9987 | −24.45 | ||||||||
| 310 | 827.18 | 1.13 | 0.9994 | −35.12 | 13.49 | 0.89 | 0.9991 | −24.50 | ||||||||
| 0.04 | 293 | 801.73 | 1.14 | 0.9999 | −33.10 | −17.81 | 52.21 | 0.9995 | 18.98 | 0.92 | 0.9969 | −23.96 | −18.95 | 17.11 | 0.9959 | |
| 298 | 700.91 | 1.14 | 0.9991 | −33.36 | 16.25 | 0.92 | 0.9989 | −24.05 | ||||||||
| 304 | 610.89 | 1.14 | 0.9998 | −33.68 | 13.88 | 0.92 | 0.9975 | −24.15 | ||||||||
| 310 | 535.97 | 1.14 | 0.9998 | −33.99 | 12.40 | 0.92 | 0.9980 | −24.25 | ||||||||
| 0.09 | 293 | 531.20 | 1.13 | 0.9996 | −32.08 | −28.48 | 12.29 | 0.9987 | 17.55 | 0.93 | 0.9973 | −23.82 | −21.75 | 7.08 | 0.9978 | |
| 298 | 422.46 | 1.12 | 0.9992 | −32.14 | 15.18 | 0.93 | 0.9946 | −23.86 | ||||||||
| 304 | 344.16 | 1.12 | 0.9991 | −32.22 | 13.04 | 0.93 | 0.9975 | −23.90 | ||||||||
| 310 | 277.44 | 1.12 | 0.9992 | −32.29 | 10.68 | 0.91 | 0.9944 | −23.94 | ||||||||
| 0.15 | 293 | 343.36 | 1.11 | 0.9997 | −31.02 | −25.87 | 17.56 | 0.9972 | 15.36 | 0.93 | 0.9966 | −23.42 | −23.12 | 1.03 | 0.9940 | |
| 298 | 280.86 | 1.11 | 0.9994 | −31.10 | 12.38 | 0.92 | 0.9976 | −23.43 | ||||||||
| 304 | 225.54 | 1.11 | 0.9996 | −31.21 | 10.58 | 0.92 | 0.9968 | −23.44 | ||||||||
| 310 | 192.48 | 1.11 | 0.9967 | −31.32 | 9.03 | 0.92 | 0.9990 | −23.44 | ||||||||
| BSA- AG50 | 0 | 293 | 115.49 | 0.95 | 0.9985 | −28.40 | −12.11 | 55.58 | 0.9999 | 16.29 | 0.88 | 0.9990 | −23.61 | −13.63 | 34.06 | 0.9919 |
| 298 | 106.23 | 0.96 | 0.9990 | −28.68 | 14.74 | 0.88 | 0.9990 | −23.78 | ||||||||
| 304 | 96.79 | 0.97 | 0.9999 | −29.01 | 12.89 | 0.89 | 0.9966 | −23.98 | ||||||||
| 310 | 87.82 | 0.97 | 0.9988 | −29.34 | 12.08 | 0.90 | 0.9994 | −24.19 | ||||||||
| 0.04 | 293 | 100.63 | 0.98 | 0.9996 | −28.05 | −12.94 | 51.55 | 0.9992 | 15.39 | 0.91 | 0.9977 | −23.46 | −16.29 | 24.47 | 0.9952 | |
| 298 | 90.98 | 0.99 | 0.9995 | −28.31 | 13.47 | 0.91 | 0.9996 | −23.58 | ||||||||
| 304 | 82.63 | 1.00 | 0.9982 | −28.62 | 11.73 | 0.90 | 0.9975 | −23.73 | ||||||||
| 310 | 74.99 | 1.00 | 0.9992 | −28.93 | 10.69 | 0.90 | 0.9974 | −23.87 | ||||||||
| 0.09 | 293 | 89.19 | 1.00 | 0.9999 | −27.76 | −11.52 | 55.42 | 0.9997 | 14.56 | 0.91 | 0.9986 | −23.33 | −19.38 | 13.51 | 0.9989 | |
| 298 | 82.05 | 1.00 | 0.9989 | −28.04 | 12.57 | 0.91 | 0.9945 | −23.40 | ||||||||
| 304 | 74.77 | 1.01 | 0.9999 | −28.37 | 10.74 | 0.91 | 0.9950 | −23.48 | ||||||||
| 310 | 68.83 | 1.02 | 0.9988 | −28.70 | 9.42 | 0.90 | 0.9928 | −23.56 | ||||||||
| 0.15 | 293 | 80.63 | 1.02 | 0.9994 | −27.50 | −14.14 | 45.59 | 0.9947 | 13.71 | 0.95 | 0.9982 | −23.20 | −17.53 | 19.33 | 0.9985 | |
| 298 | 70.97 | 1.02 | 0.9986 | −27.72 | 11.99 | 0.95 | 0.9983 | −23.29 | ||||||||
| 304 | 65.34 | 1.03 | 0.9995 | −28.00 | 10.65 | 0.94 | 0.9982 | −23.41 | ||||||||
| 310 | 58.01 | 1.03 | 0.9992 | −28.27 | 9.17 | 0.94 | 0.9957 | −23.53 | ||||||||
Ra and Rb are the correlation coefficients for K values and Van’t Hoff plots, respectively.
Acknowledgements
The authors gratefully acknowledge financial support from Sichuan Provincial Science & Technology Fund for Applied Fundamental Research (2016JY0080), Open Fund (CSPC2013-6) of Chemical Synthesis and Pollution Control Key Laboratory of Sichuan Province (China West Normal University), and the Education Department of Sichuan Province (12ZA171).
Footnotes
Transparency data associated with this article can be found in the online version at 10.1016/j.dib.2016.06.025.
Contributor Information
Zhengjun Cheng, Email: ncczj1112@126.com.
Lei Fan, Email: fanlei@yzu.edu.cn.
Transparency document. Supplementary material
Supplementary material
References
- 1.Li T., Cheng Z.J., Cao L.J., Jiang X.H. Comparison of interactions between three food colorants and BSA. Food Chem. 2016;194:740–748. doi: 10.1016/j.foodchem.2015.08.067. [DOI] [PubMed] [Google Scholar]
- 2.Cheng Z.J., Liu R., Jiang X.H. Spectroscopic studies on the interaction between tetrandrine and two serum albumins by chemometrics methods. Spectrochim. Acta Part A-Mol. Biomol. Spectrosc. 2013;115:92–105. doi: 10.1016/j.saa.2013.06.007. [DOI] [PubMed] [Google Scholar]
- 3.Bolel P., Datta S., Mahapatra N., Halder M. Exploration of pH-dependent behavior of the anion receptor pocket of subdomain IIA of HSA: determination of effective pocket charge using the Debye–Hückel limiting law. J. Phys. Chem. B. 2014;118:26–36. doi: 10.1021/jp407057f. [DOI] [PubMed] [Google Scholar]
- 4.Shahabadi N., Maghsudi M., Kiani Z., Pourfoulad M. Multispectroscopic studies on the interaction of 2-tert-butylhydroquinone (TBHQ), a food additive, with bovine serum albumin. Food Chem. 2011;124:1063–1068. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material