Abstract
BACKGROUND
The EphA2 receptor tyrosine kinase is known to promote cancer cell malignancy in the absence of activation by ephrin ligands. This behavior depends on high EphA2 phosphorylation on Ser897 and low tyrosine phosphorylation, resulting in increased cell migration and invasiveness. We have previously shown that EphA2 forms dimers in the absence of ephrin ligand binding, and that dimerization of unliganded EphA2 can decrease EphA2 Ser897 phosphorylation. We have also identified a small peptide called YSA, which binds EphA2 and competes with the naturally occurring ephrin ligands.
METHODS
Here, we investigate the effect of YSA on EphA2 dimer stability and EphA2 function using quantitative FRET techniques, Western blotting, and cell motility assays.
RESULTS
We find that the YSA peptide stabilizes the EphA2 dimer, increases EphA2 Tyr phosphorylation, and decreases both Ser897 phosphorylation and cell migration.
CONCLUSIONS
The experiments demonstrate that the small peptide ligand YSA reduces EphA2 Ser897 pro-tumorigenic signaling by stabilizing the EphA2 dimer.
GENERAL SIGNIFICANCE
This work is a proof-of-principle demonstration that EphA2 homointeractions in the plasma membrane can be pharmacologically modulated to decrease the pro-tumorigenic signaling of the receptor.
INTRODUCTION
RTKs are a large superfamily of 58 receptors, divided into 20 families (1-3). They are single-pass transmembrane proteins with an extracellular ligand-binding region, a transmembrane segment and an intracellular catalytic domain. Dimerization is required for RTK activation, as the proximity of two kinase domains in the dimer results in cross-phosphorylation of receptor molecules on selected tyrosines as the first step of the activation process. According to the canonical model of RTK activation, dimerization is induced by ligand binding (1). However, recent work has demonstrated that some RTKs have intrinsic propensity for dimerization, and thus they can become phosphorylated on tyrosine residues and activated even in the context of unliganded dimers (4-6). As RTK dimerization obeys the law of mass action, high receptor expression leads to higher dimeric fraction and, therefore, to higher activity (6, 7). Thus, enhanced RTK unliganded dimerization can contribute to tumorigenesis and cancer progression in malignant cells that overexpress oncogenic RTKs (8-15).
The Eph receptor family is the largest of the 20 RTK families. The Eph receptors regulate axon guidance, bone morphogenesis, and angiogenesis as well as play important roles in inflammatory responses, neurodegenerative diseases, and cancer (16, 17). These receptors are known to form large oligomers upon binding to their ligands (ephrins), which are anchored on the surface of opposing cells (18-22). In response to ligand binding, the Eph receptors cross-phosphorylate each other on two tyrosines in the juxtamembrane domain and a tyrosine in the activation loop. This often results in cell contraction and disruption of cell-cell contacts (23-26), ultimately leading to inhibition of cell migration and invasiveness (16, 27-29).
A member of the Eph receptor family, EphA2, can promote cell migration/invasiveness and tumor malignancy in a ligand-independent manner (30). This tumorigenic activity has been linked to high levels of phosphorylation on Ser897 in the linker segment between the kinase domain and the SAM domain and to low levels of tyrosine phosphorylation (30-33). Thus, EphA2 appears to behave as an oncoprotein in the absence of ligand binding, which may explain why its overexpression is associated with poor prognosis in many cancers (34-37). Consistent with this view, EphA2 overexpression in cancer is often accompanied by the loss of ephrin ligands (16, 38).
While EphA2 was believed to be monomeric when not bound to ephrin ligands, we have recently demonstrated that this receptor can form dimers in the plasma membrane of HEK293T cells in the absence of ephrin ligand binding (39). Furthermore, we have shown that mutation to Arg of three hydrophobic amino acids in the extracellular cysteine-rich region (L223, L254, and V255) destabilizes the EphA2 dimer. The dimerization-deficient EphA2 mutant shows enhanced ability to promote cell migration concomitant with increased phosphorylation on Ser897 and decreased tyrosine phosphorylation as compared to EphA2 wild-type (39). These findings suggest that the EphA2 monomer favors pro-tumorigenic signaling. Therefore, unique EphA2-specific therapeutic strategies may be needed for cancers driven by EphA2 overexpression. In particular, molecules that stabilize EphA2 dimers will reduce EphA2 monomer concentration as well as promote the tumor suppressor activity associated with EphA2 kinase-dependent signaling (16, 30).
Using phage display, we previously identified two dodecameric peptides that bind selectively to EphA2 with low micromolar affinity (40, 41). These peptides interact with the ephrin-binding pocket in the ligand-binding domain of EphA2 and compete with ephrin ligands for binding (40, 42). Computer modeling of peptide binding to the ephrin-binding pocket of EphA2 and structure-activity relationship studies have revealed a 1:1 binding stoichiometry (42, 43), which has been supported by ITC data (44, 45).
Intriguingly, these peptides mimic the ephrins by acting as agonists that increase EphA2 tyrosine phosphorylation (40, 42-44, 46) and downstream signaling (42, 46). The mechanism of EphA2 activation induced by these small peptides is currently unknown. Here we show that one of these peptides, referred to as “YSA”, may inhibit the tumorigenic activity of EphA2 by stabilizing the dimeric form of the receptor.
MATERIALS AND METHODS
Cell culture and transfection
The production of the pcDNA 3.1(+) EphA2-15aa-mTurq and pcDNA3.1(+) EphA2-15aa-eYFP constructs has been described previously (39). These constructs encode full-length EphA2, followed by a flexible linker consisting of 5 GGS repeats, and fluorescent proteins (mTurqouise and eYFP, a FRET pair) at the C-terminus. HEK293T cells were purchased from the American Type Culture Collection (ATCC) (Manassas, VA, USA). For imaging, the cells were cultures in 35 mm glass bottom dishes (MatTek Corporation, MA) in Dulbecco's modified Eagle medium (DMEM), supplemented with 10% fetal bovine serum (FBS, Hyclone), 3.5g/L D-glucose (19.4mM) and 1.5g/L (17.9mM) sodium bicarbonate. Cells were co-transfected with pcDNA3.1 (+) EphA2-15aa-mTurq and pcDNA3.1 (+) EphA2-15aa-eYFP using Lipofectamine 2000 (Invitrogen), following the manufacturer's protocol. Twelve hours following transfection, the cells were serum-starved for 12 hours. Just before imaging, the starvation medium was replaced with hypo-osmotic medium to swell the cells under reversible conditions as described (47). The medium contained 6 μM YSA peptide.
Two photon microscopy of cells under reversible osmotic stress
The swollen cells in the presence of the YSA peptide were imaged in a spectrally resolved two photon microscope with line-scanning capabilities. A mode-locked laser (MaiTai™, Spectra-Physics, Santa Clara) that generates femtosecond pulses between wavelengths 690 nm to 1040 nm was used to excite the fluorophores mTurquoise and eYFP. Details about the microscope are given in previous publications (48, 49). Measurements were performed in cells under reversible osmotic stress. The swelling step was necessary because plasma membranes are very “wrinkled”, preventing determination of exact two-dimensional receptor concentrations. When the membrane is stretched, however, a calibration with purified fluorescent protein solutions of known concentrations can be used to calculate the receptor concentrations per unit membrane area (29).
The transfected cells were cultured under starvation conditions prior to the experiments to ensure that no soluble ligand is present. Furthermore, the cell culture medium was replaced just before imaging, thereby removing any traces of ephrins that may be present in the solution. We imaged only plasma membranes of swollen cells that were not in contact with neighboring cells, ensuring that the EphA2 receptors did not interact with ephrins that may be present on opposing cells.
Measurements of dimerization propensities
EphA2 dimerization propensities were measured using the Fully Quantified Spectral Imaging FRET (FSI-FRET) method. The receptor concentrations were varied over a wide range, and the concentrations in the plasma membranes were measured, along with the FRET efficiencies. The total receptor concentration and the dimeric receptor fraction were calculated for small membrane segments (Figure 1), and data from many cells were combined to obtain dimerization curves. From the FRET data, two parameters were determined to describe the EphA2 dimerization process. The first parameter is the dimer stability, ΔG, which is linked to the dissociation constant Kdiss (in units of receptors/μm2). The standard state for ΔG is defined as nm2/receptor (50), and therefore:
| (1) |
The second parameter determined from the fit is the structural parameter “Intrinsic FRET”, Ẽ, which depends primarily on the distance d between the fluorescent proteins in the dimer according to:
| (2) |
Here Ro is the Förster radius of the mTurq/eYFP FRET pair, 54.5 Å.
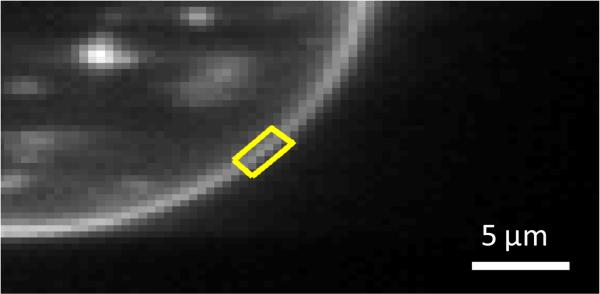
Figure 1.
Selection of a membrane region for FSI-FRET analysis. The cell under reversible osmotic stress exhibits large areas of stretched membrane with homogeneously distributed EphA2 fluorescence. Each data point in Figures 2 and 4 corresponds to a homogeneous region in a cell membrane, ~3 μm in length (outlined in yellow).
Western blots
Twenty four hours following transfection, the cells were incubated with medium containing 6 μM of YSA. The cells were placed in an incubator for 10 minutes and then treated with lysis buffer (25mM Tris-Cl, 0.5% TritonX-100, 20mM NaCl, 2mM EDTA, phosphate and protease inhibitors (Roche Applied Science)). The lysed samples were centrifuged at 14,000 g for 15 minutes at 4°C and stored at −20°C. The amounts of total protein in the lysates were measured with the BCA protein assay kit (Bio-Rad, CA). The lysates were loaded into 3–8% NuPAGEHNovexHTris-Acetate mini gels (Invitrogen, CA). The proteins were transferred onto a nitrocellulose membrane, and blocked using 5% non-fat milk in 1×TBST. Total EphA2 expression, Ser897 phosphorylation and Tyr772 phosphorylation were probed using anti-EphA2 antibodies (Cell Signalling, MA), anti-phospho-Ser897 antibodies (Cell Signaling, MA), and anti-phospho-Tyr772 antibodies (Cell Signaling, MA), respectively. Anti-rabbit HRP conjugated antibodies (Promega) were used as secondary antibodies to visualize the EphA2 expression and phosphorylation bands. The membranes were incubated with Amersham ECL Plus™ Western Blotting Detection Reagent (GE HealthCare Life Sciences, PA) for 2 minutes and then exposed for 1 to 60 seconds in a Chemidoc molecular imager (Bio-Rad, CA) to detect the chemiluminescent bands.
Cell migration assay
HEK293T cells were cultured for 24 hours following transfection and then serum starved overnight. A cell suspension with 1×106 cells per ml was prepared in serum free medium containing 0.5% BSA. The CytoSelect™ Cell Haptotaxis Assay Kit (CellBiolabs, CA) was used to assay the migratory properties of cells. This kit contains inserts with polycarbonate membrane with pores of size 8μm, coated with collagen I on the bottom side. Five hundred μl of medium containing 10% FBS were placed in the lower well. The inserts were loaded with 300 μl of the cell suspension and the YSA peptide was added to the cells at a final concentration of 6 μM. The cells were allowed to migrate for 4 hours at 37°C. After that, the medium was aspirated from the inserts, and the top of the polycarbonate membrane was cleaned using cotton swabs to remove non-migratory cells. The inserts were then transferred to clean wells containing 300 μl of Lysis Buffer/CyQuant® GR dye solution, supplied with the kit. After a 10 minute incubation at room temperature, 200 μl of the lysates were transferred to a well in a 96 well plate. The florescence of the CyQuant® GR dye solution, which is directly proportional to the number of cells that had migrated to the bottom side of the polycarbonate membrane, was measured in a plate reader.
RESULTS
The YSA peptide (YSAYPDSVPMMS) binds specifically to EphA2(41). Prior work with this peptide utilized a biotinylated variant containing a C-terminal GSGSK linker through which biotin was attached(40). The biotinylated YSA peptide has a higher antagonistic potency than the unmodified YSA, with a dissociation constant (KD) of ~200 nM measured by surface plasmon resonance(40),(41). Thus, we have used the YSA peptide with the GSGSK linker (YSAYPDSVPMMSGSGSK).
EphA2 dimerization propensity on the surface of live HEK293T cells was measured using FRET in the presence of 6 μM YSA peptide in the culture medium. At this concentration, YSA is expected to exceed both the YSA-EphA2 dissociation constant and the concentration of the EphA2 receptor. Thus, a large population (likely nearly all) of the EphA2 receptors is expected to be bound to YSA during our imaging experiments.
The FRET data were acquired in a two-photon microscope with spectral acquisition capabilities using the Fully Quantified Spectral Imaging (FSI)-FRET technique/method that has been published previously(51). This method yields (i) the donor concentration, (ii) the acceptor concentration, and (iii) the FRET efficiencies in select regions of stretched plasma membrane not in contact with other cells (as illustrated in Figure 1).
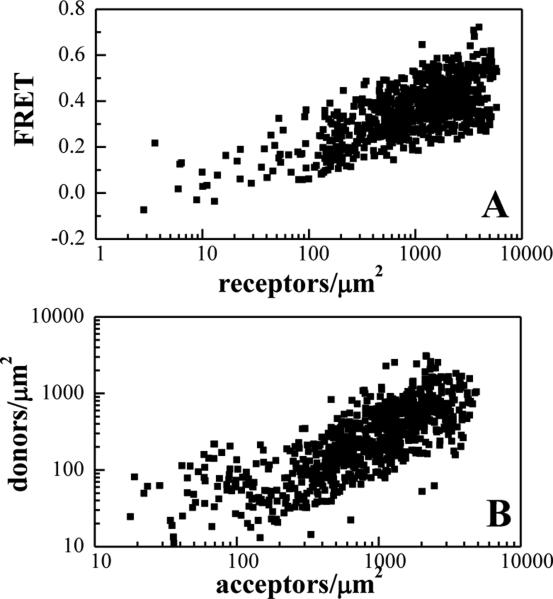
The FRET data for EphA2 in the presence of YSA, acquired from 320 cells expressing different levels of EphA2, are shown in Figure 2. Each data point corresponds to a small area of stretched cell membrane under reversible osmotic stress. Figure 2A shows the FRET efficiencies as a function of receptor (donor + acceptor) concentration. The high FRET efficiencies are indicative of specific interactions between the YSA-bound EphA2 receptors. Figure 2B shows the donor versus the acceptor concentration, which were varied over a very broad range in these transient transfection experiments. As discussed previously (51, 52), such FRET data are necessary and sufficient to characterize the lateral interactions in the membrane in quantitative terms.
Figure 2. FRET efficiencies and EphA2 concentrations measured for the YSA-bound receptor.
(A) FRET as a function of receptor concentration. Every data point represents a single membrane region such as the one shown in Figure 1. (B) Receptor donor concentration plotted as a function of receptor acceptor concentration in each membrane region analyzed.
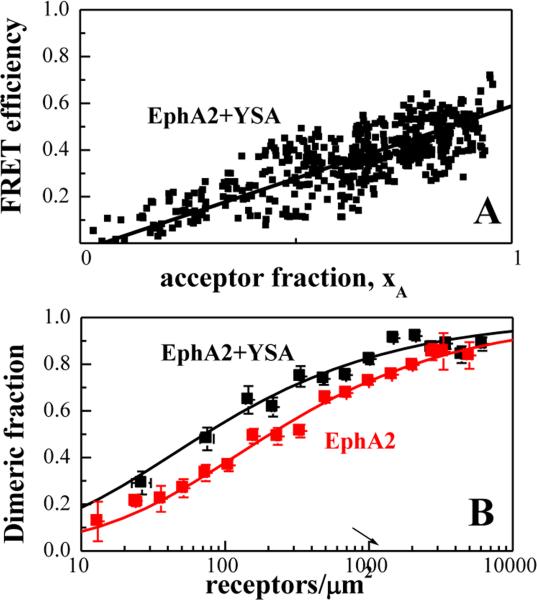
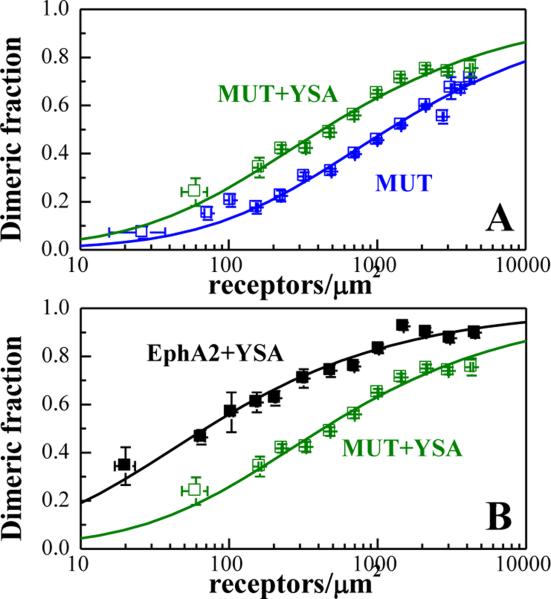
To evaluate whether EphA2 bound to YSA forms dimers or higher order oligomers, in Figure 3A we plotted the FRET efficiency as a function of acceptor fraction, xA, under conditions where FRET does not strongly depend on receptor concentration. In particular, we selected data points corresponding to membrane regions in which EphA2 concentrations exceeded 2,000 receptors/μm2 because FRET does not vary substantially with concentration at these high levels of expression (Figure 2A). Under such conditions, the dependence of FRET efficiency on the acceptor fraction is known to be linear for a dimer and non-linear for higher order oligomers (53-55). The data in Figure 3A are well described by a linear function, demonstrating that YSA-bound EphA2 forms dimers.
Figure 3. The YSA peptide promotes EphA2 dimerization.
(A) FRET efficiency as a function of receptor acceptor fraction, for total EphA2 concentrations that exceed 2,000 receptors/μm2. Under these conditions, the FRET efficiency depends primarily on the acceptor fraction (xA) and the linear dependence suggests that the oligomer type is a dimer(53-55). (B) Dimerization curves for YSA-bound EphA2 are compared to previously published dimerization curves for unliganded EphA2(39). The dimeric fractions are binned, and averages and standard errors are shown for each bin. The solid line, given by , is the theoretical curve for the best-fit dimerization model. YSA binding stabilizes the EphA2 dimer by −0.7 ± 0.3 kcal/mole.
Next, the FRET data for YSA-bound EphA2 were fitted to a dimerization model with two adjustable parameters as previously described (39, 51). The first parameter is the dissociation constant, Kdiss, which is used to calculate the dimerization free energy ΔG indicative of dimer stability. The second parameter is the structural parameter “Intrinsic FRET”, Ẽ, which depends on the dimer structure, and in particular on the average distance and orientation of the fluorescent proteins in the dimer (50). Since the fluorescent proteins are attached via long flexible linkers, Intrinsic FRET is expected to depend primarily on the distance between the fluorescent proteins. The optimal parameters, determined in the fit, are shown in Table 1.
Table 1.
Dissociation constants and dimerization free energies for YSA-bound EphA2 are compared to published values for unliganded EphA2(39).
| Kdiss (rec/μm2) | ΔG(kcal/mole) | Ẽ | d (Å) | |
|---|---|---|---|---|
| EphA2** | 210 ± 50 | −5.0 ± 0.20 | 0.67 ± 0.03 | 48 ± 1 |
| EphA2 + YSA | 69 ± 23 | −5.7 ± 0.24 | 0.52 ± 0.02 | 54 ± 1 |
| Mutant* EphA2** | 1200 ± 250 | −4.0 ± 0.20 | 0.72 ± 0.04 | 47 ± 1 |
| Mutant* + YSA | 428 ± 103 | −4.6 ± 0.16 | 0.54 ± 0.03 | 53 ± 1 |
Also shown are the Intrinsic FRET values, Ẽ, from which averages distances between the two fluorescent proteins in the dimer, d, are calculated.
L223R/L254R/V255R EphA2 mutant.
Data from (39).
The best-fit dimerization curve for EphA2 in the presence of YSA is shown in Figure 3B (black symbols) and compared to the previously published dimerization curve for EphA2 in the absence of YSA (red symbols). Both unliganded EphA2 and YSA-bound EphA2 form dimers, but the YSA-bound EphA2 dimer is more stable by −0.7 ± 0.3 kcal/mole (Table 1, ΔG). Thus, YSA binding stabilizes the EphA2 dimers. The Intrinsic FRET of EphA2 also changes upon YSA binding (Table 1), indicative of a structural change in the EphA2 dimer upon YSA binding. This structural change involves an increase in fluorescent protein separation in the dimer.
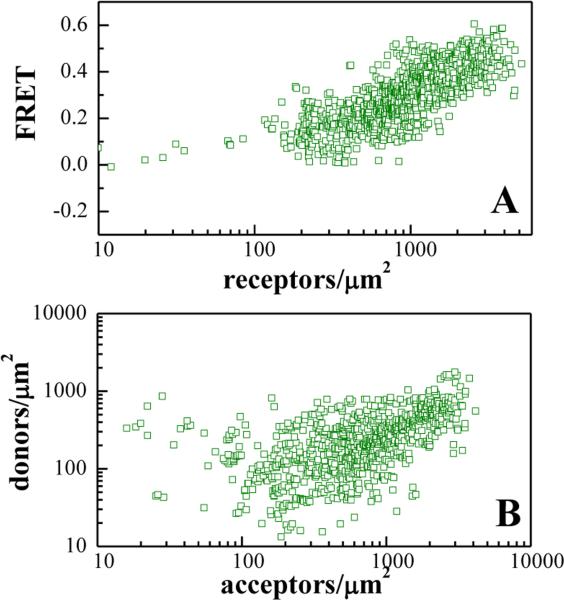
Next we asked whether the observed change in Intrinsic FRET is indicative of a YSA-induced switch to a distinctly different EphA2 dimer architecture with a different receptor-receptor contact interfaces. Alternatively, only modest allosteric structural changes may be occurring in the EphA2 dimer upon YSA binding, without alterations in the contact interfaces. We specifically focused on a receptor-receptor interface involving L223, L254, and V255 in the cysteine-rich region of the extracellular domain, which has previously been shown to stabilize EphA2 dimers in the absence of ligand binding (39). We therefore performed FRET experiments with the EphA2 L223R/L254R/V255R mutant in the presence of 6 μM YSA. The FRET data are shown in Figure 4, and the dimerization curve is shown in Figure 5 with the green symbols. This dimerization curve is compared to the dimerization curve for the EphA2 L223R/L254R/V255R mutant in the absence of YSA (Figure 5A) and to the dimerization curve for wild-type EphA2 in the presence of YSA (Figure 5B). In Figure 5A, we see that YSA binding stabilizes the L223R/L254R/V255R EphA2 mutant dimer. In Figure 5B, we see that the L223R/L254R/V255R mutations destabilize the YSA-bound EphA2 dimer. The reduction in YSA-bound EphA2 dimer stability due to the L223R/L254R/V255R mutations is 1.1 ± 0.3 kcal/mole. This destabilizing effect is very similar to the effect of the L223R/L254R/V255R mutation on unliganded EphA2 dimer stability (1.0 ± 0.3 kcal/mole) (39). These data demonstrate that contacts mediated by amino acids L223, L254, and V255 stabilize EphA2 dimers both in the presence and absence of the YSA peptide. Thus, YSA binding to the EphA2 dimer seems to preserve the interface in the cysteine-rich region, where the mutated amino acids are located.
Figure 4. FRET efficiencies and concentrations measured for the YSA-bound EphA2 L223R/L254R/V255R mutant.
(A) FRET as a function of receptor concentration. Every data point represents a single membrane region such as the one shown in Figure 1. (B) Receptor donor concentration plotted as a function of receptor acceptor concentration in each membrane region analyzed.
Figure 5. The YSA peptide promotes dimerization of the EphA2 L223R/L254R/V255R mutant.
(A) Dimerization curves for the YSA-bound EphA2 mutant are compared to previously published dimerization curves for the unliganded mutant.(39) (B) Comparison of the dimerization curves for YSA-bound EphA2 wild-type and mutant.
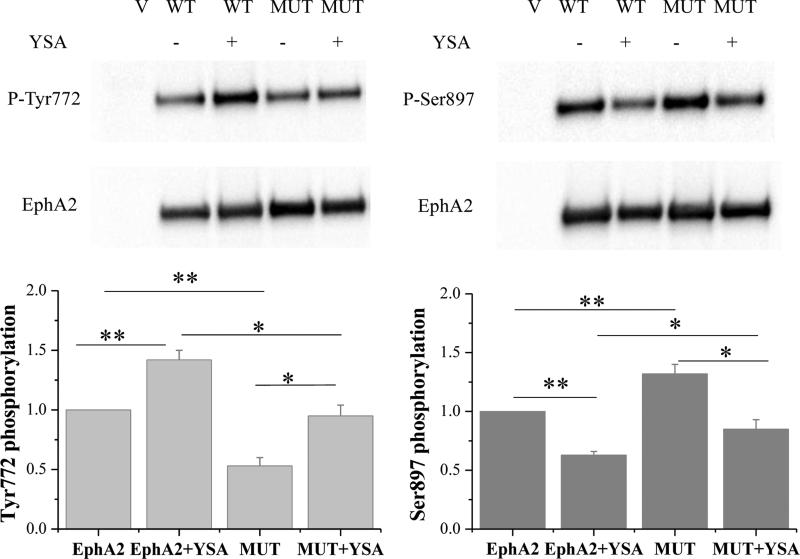
We also found that the YSA peptide at a concentration of 6 μM increases by ~40% the tyrosine phosphorylation of both wild-type EphA2 and the EphA2 L223R/L254R/V255R mutant (p = 0.01 for both forms of the receptor; Figure 6). Similarly, YSA decreased the Ser897 phosphorylation of EphA2 wild-type by ~40% and that of the EphA2 L223R/L254R/V255R mutant by ~35% (p<0.001 for both forms of the receptor; Figure 6). Furthermore, the L223R/L254R/V255R mutations increased EphA2 Ser897 phosphorylation and decreased EphA2 tyrosine phosphorylation, supporting the conclusion that these mutations destabilize not only the unliganded EphA2 dimer but also the YSA-induced EphA2 dimer.
Figure 6. YSA binding increases EphA2 Tyr772 phosphorylation and reduces Ser897 phosphorylation.
Top: Representative Western blots. Bottom: Quantification from three independent experiments. Shown are means and standard errors. *, p < 0.05; **, p < 0.01.
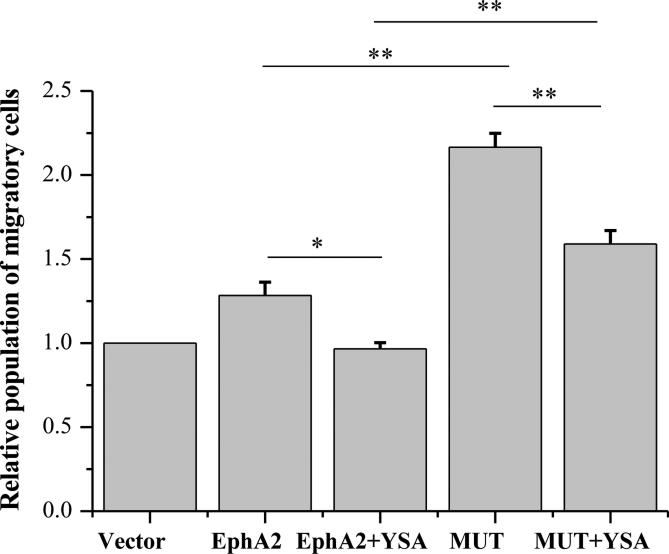
Finally, in transwell cell migration assays, the YSA peptide suppressed the migratory ability of HEK293T cells by ~25% for both EphA2 wild-type (p < 0.05) and mutant (p < 0.01) (Figure 7). This demonstrates that the YSA peptide can inhibit the pro-migratory effects of EphA2. Furthermore, the L223R/L254R/V255R mutation increased the migratory potential of the HEK293T cells by ~50 % in the presence of the YSA peptide, consistent with the hypothesis that the mutated residues participate in EphA2 wild-type dimerization upon YSA binding.
Figure 7. YSA binding to EphA2 reduces cell migration.
Shown are means and standard errors from three independent experiments with HEK293T cells. *: p < 0.05; **: p < 0.01.
Discussion
The YSA peptide stabilizes the EphA2 dimer
Using the FSI-FRET method, we previously showed that EphA2 can form dimers on the cell surface in the absence of ephrin ligand binding (39). Based on prior structural data and our own work, we now have an understanding of the general architecture of this dimer. First, we have shown that L223, L254, and V255 in the cysteine-rich domain play a role in EphA2 dimer stabilization (39). These contacts are part of an interface that does not involve the ephrin ligands and was thus designated “clustering interface” (19). This clustering interface involves lateral contacts between both the cysteine-rich domain domains and the ligand-binding domains (18, 19). Second, NMR structures of the isolated EphA2 transmembrane segments in detergent micelles, molecular modeling, and activity studies have suggested that specific receptor-receptor contacts occur between the transmembrane segments in the absence of ligand(56). Third, it can be expected that contacts occur between the kinase domains, as such contacts are likely required for the EphA2 tyrosine phosphorylation that accompanies the dimerization of unliganded EphA2 in transiently transfected HEK293T cells. In fact, contacts between the two EphA2 molecules in the dimer likely occur along the length of the receptors (57).
Although it inhibits ephrin binding, the YSA peptide has been shown to function as an EphA2 agonist (40, 41). Furthermore, molecular docking simulations have suggested that the YSA peptide occupies the ephrin-binding pocket of EphA2 (42). The YSA binding site, therefore, is likely to be on the opposite side of the so-called “clustering interface” in the extracellular region, which is in part formed by amino acids L223, L254, and V255(19).
It has been previously hypothesized that YSA binding causes conformational changes in the EphA2 ligand-binding domain that propagate to other domains of the receptor (41). Our finding that YSA binding stabilizes the EphA2 dimer supports this hypothesis, and suggests that the YSA peptide exerts its dimer stabilization effect via an allosteric mechanism. In other words, YSA binding leads to alterations in the EphA2 dimer interface, and these alterations stabilize the EphA2 dimer. The inferred structural changes are propagated along the entire length of the EphA2 dimer, and can be detected as a change in Intrinsic FRET following YSA binding.
Currently, it is not known in detail how the EphA2 dimer interface along the entire length of the receptor changes upon YSA binding. In our mutagenesis experiments, we only focus on a specific portion of the CRD domain and we show that the YSA-free and YSA-bound dimers are stabilized through contacts involving the same three amino acids, L223, L254R, and V255R. This conclusion stems from our finding that the L223R/L254R/V255R mutations destabilize the YSA-bound EphA2 dimer to the same extent as they destabilize the unliganded EphA2 dimer. This finding, however, does not exclude the possibility that subtle structural changes occur even in the CRD domain upon YSA binding.
Implications
The EphA2 receptor tyrosine kinase promotes cancer cell malignancy via a ligand-independent process that is unique among the RTKs and involves phosphorylation on Ser897. Often EphA2 Ser897 phosphorylation correlates with low receptor tyrosine phosphorylation and increased cell migratory potential (30). This behavior of EphA2 is also manifested in transiently transfected HEK293T cells (30). We recently showed that ligand-independent EphA2 dimerization in HEK293T cells correlates with high receptor tyrosine phosphorylation, low phosphorylation on Ser897 and decreased cell migratory potential (39), thus establishing the biological significance of EphA2 homo-interactions in the absence of ligand.
Unliganded dimers for other RTKs are known to form, but their activity is rather low (referred to as “basal activity”) (6, 58, 59). Ligand binding to these preformed dimers has been shown to change the dimer structure, and increase the activity (6, 58, 59). Thus, unliganded RTK dimers typically play roles of signaling intermediates. On the other hand, EphA2 dimers play a unique role in inhibiting pro-oncogenic signaling by depleting EphA2 monomer populations.
The mechanism of EphA2-mediated tumorigenesis in the absence of ligand therefore appears to be the opposite to the tumorigenesis mechanism of a typical RTK (such as EGFR, FGFR, etc). In other words, unliganded dimerization is pro-tumorigenic for most RTKs (8, 9, 11, 15, 60-64), but it is anti-tumorigenic for EphA2. Therefore novel, distinctly different therapeutic strategies may be needed for cancers that arise due to EphA2 overexpression (16, 29). For instance, inhibitors of receptor kinase activity similar to those that are currently being developed for other RTKs may actually enhance, instead of decreasing, EphA2 pro-tumorigenic signaling. Our work suggests that one possible EphA2-specific therapeutic strategy may be to minimize EphA2 monomer populations by stabilizing EphA2 dimers or oligomers.
The YSA-induced increase in EphA2 stability correlates with an increase in tyrosine phosphorylation (indicative of kinase activity) and a decrease in Ser897 phosphorylation as well as an decrease in cell migration. These results are a direct demonstration that a small peptide can reduce cell migration via a mechanism that involves stabilization of EphA2 dimers. In particular, the peptide exerts control over EphA2 tumorigenic signaling ability by modulating receptor homo-interactions in the plasma membrane. Peptides with this property could be potentially developed into EphA2-specific therapies with utility in the clinic.
Highlights.
Small peptide ligands are known to increase EphA2 tyrosine phosphorylation
We show that the YSA peptide ligand stabilizes the EphA2 dimer
The YSA peptide ligand reduces EphA2 Ser897 phosphorylation
The YSA peptide ligand decreases cell migration
ACKNOWLEDGEMENT
Supported by NSF MCB 1157687 and NIH GM068619 (to K.H.) and CA138390 (to E.B.P.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- 1.Fantl WJ, Johnson DE, Williams LT. Signaling by Receptor Tyrosine Kinases. Annu. Rev. Biochem. 1993;62:453–481. doi: 10.1146/annurev.bi.62.070193.002321. [DOI] [PubMed] [Google Scholar]
- 2.Lemmon MA, Schlessinger J. Cell Signaling by Receptor Tyrosine Kinases. Cell. 2010;141:1117–1134. doi: 10.1016/j.cell.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2000;103:211–225. doi: 10.1016/s0092-8674(00)00114-8. [DOI] [PubMed] [Google Scholar]
- 4.Low-Nam ST, Lidke KA, Cutler PJ, Roovers RC, van Bergen en Henegouwen PM, Wilson BS, Lidke DS. ErbB1 dimerization is promoted by domain co- confinement and stabilized by ligand binding. Nat. Struct. Mol. Biol. 2011;18:1244–1249. doi: 10.1038/nsmb.2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Endres NF, Das R, Smith AW, Arkhipov A, Kovacs E, Huang YJ, Pelton JG, Shan YB, Shaw DE, Wemmer DE, Groves JT, Kuriyan J. Conformational Coupling across the Plasma Membrane in Activation of the EGF Receptor. Cell. 2013;152:543–556. doi: 10.1016/j.cell.2012.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sarabipour S, Hristova K. Mechanism of FGF receptor dimerization and activation. Nat. Commun. 2016;7:10262. doi: 10.1038/ncomms10262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.He L, Hristova K. Physical-chemical principles underlying RTK activation, and their implications for human disease. Biochim. Biophys. Acta. 2012;1818:995–1005. doi: 10.1016/j.bbamem.2011.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barker FG, Simmons ML, Chang SM, Prados MD, Larson DA, Sneed PK, Wara WM, Berger MS, Chen PC, Israel MA, Aldape KD. EGFR overexpression and radiation response in glioblastoma multiforme. International Journal of Radiation Oncology Biology Physics. 2001;51:410–418. doi: 10.1016/s0360-3016(01)01609-1. [DOI] [PubMed] [Google Scholar]
- 9.Browne BC, O'Brien N, Duffy MJ, Crown J, O'Donovan N. HER-2 Signaling and Inhibition in Breast Cancer. Current Cancer Drug Targets. 2009;9:419–438. doi: 10.2174/156800909788166484. [DOI] [PubMed] [Google Scholar]
- 10.Brunelleschi S, Penengo L, Santoro MM, Gaudino G. Receptor tyrosine kinases as target for anti-cancer therapy. Current Pharmaceutical Design. 2002;8:1959–1972. doi: 10.2174/1381612023393530. [DOI] [PubMed] [Google Scholar]
- 11.Chesi M, Nardini E, Lim RSC, Smith KD, Kuehl WM, Bergsagel PL. The t(4;14) translocation in myeloma dysregulates both FGFR3 and a novel gene, MMSET, resulting in IgH/MMSET hybrid transcripts. Blood. 1998;92:3025–3034. [PubMed] [Google Scholar]
- 12.Ross JS, Slodkowska EA, Symmans WF, Pusztai L, Ravdin PM, Hortobagyi GN. The HER-2 Receptor and Breast Cancer: Ten Years of Targeted Anti- HER-2 Therapy and Personalized Medicine. Oncologist. 2009;14:320–368. doi: 10.1634/theoncologist.2008-0230. [DOI] [PubMed] [Google Scholar]
- 13.Sergina NV, Moasser MM. The HER family and cancer: emerging molecular mechanisms and therapeutic targets. Trends in Molecular Medicine. 2007;13:527–534. doi: 10.1016/j.molmed.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chesi M, Brents LA, Fly SA, Bais C, Robbiani DF, Mesri E, Kuehl WM, Bergsagel PL. Activated fibroblast growth factor receptor 3 is an oncogene that contributes to tumor progression in multiple myeloma. Blood. 2001;97:729–736. doi: 10.1182/blood.v97.3.729. [DOI] [PubMed] [Google Scholar]
- 15.Turner N, Pearson A, Sharpe R, Lambros M, Geyer F, Lopez-Garcia MA, Natrajan R, Marchio C, Iorns E, Mackay A, Gillett C, Grigoriadis A, Tutt A, Reis JS, Ashworth A. FGFR1 Amplification Drives Endocrine Therapy Resistance and Is a Therapeutic Target in Breast Cancer. Cancer Research. 2010;70:2085–2094. doi: 10.1158/0008-5472.CAN-09-3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pasquale EB. Eph receptors and ephrins in cancer: bidirectional signalling and beyond. Nature Reviews Cancer. 2010;10:165–180. doi: 10.1038/nrc2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Funk SD, Yurdagul A, Albert P, Traylor JG, Jin L, Chen J, Orr AW. EphA2 Activation Promotes the Endothelial Cell Inflammatory Response A Potential Role in Atherosclerosis. Arteriosclerosis Thrombosis and Vascular Biology. 2012;32:686–U367. doi: 10.1161/ATVBAHA.111.242792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seiradake E, Harlos K, Sutton G, Aricescu AR, Jones EY. An extracellular steric seeding mechanism for Eph-ephrin signaling platform assembly. Nature Structural & Molecular Biology. 2010;17:398–U27. doi: 10.1038/nsmb.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Himanen JP, Yermekbayeva L, Janes PW, Walker JR, Xu K, Atapattu L, Rajashankar KR, Mensinga A, Lackmann M, Nikolov DB, Dhe-Paganon S. Architecture of Eph receptor clusters. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:10860–10865. doi: 10.1073/pnas.1004148107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu K, Tzvetkova-Robev D, Xu Y, Goldgur Y, Chan YP, Himanen JP, Nikolov DB. Insights into Eph receptor tyrosine kinase activation from crystal structures of the EphA4 ectodomain and its complex with ephrin-A5. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:14634–14639. doi: 10.1073/pnas.1311000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wimmer-Kleikamp SH, Nievergall E, Gegenbauer K, Adikari S, Mansour M, Yeadon T, Boyd AW, Patani NR, Lackmann M. Elevated protein tyrosine phosphatase activity provokes Eph/ephrin-facilitated adhesion of pre-B leukemia cells. Blood. 2008;112:721–732. doi: 10.1182/blood-2007-11-121681. [DOI] [PubMed] [Google Scholar]
- 22.Janes PW, Nievergall E, Lackmann M. Concepts and consequences of Eph receptor clustering. Seminars in Cell & Developmental Biology. 2012;23:43–50. doi: 10.1016/j.semcdb.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 23.Fang BW, Ireton RC, Zhuang GL, Takahashi T, Reynolds A, Chen J. Overexpression of EPHA2 receptor destabilizes adherens junctions via a RhoA- dependent mechanism. J. Cell Sci. 2008;121:358–368. doi: 10.1242/jcs.017145. [DOI] [PubMed] [Google Scholar]
- 24.Pratt RL, Kinch MS. Activation of the EphA2 tyrosine kinase stimulates the MAP/ERK kinase signaling cascade. Oncogene. 2002;21:7690–7699. doi: 10.1038/sj.onc.1205758. [DOI] [PubMed] [Google Scholar]
- 25.Wakayama Y, Miura K, Sabe H, Mochizuki N. EphrinA1-EphA2 Signal Induces Compaction and Polarization of Madin-Darby Canine Kidney Cells by Inactivating Ezrin through Negative Regulation of RhoA. J. Biol. Chem. 2011;286:44243–44253. doi: 10.1074/jbc.M111.267047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miura K, Nam JM, Kojima C, Mochizuki N, Sabe H. EphA2 Engages Git1 to Suppress Arf6 Activity Modulating Epithelial Cell-Cell Contacts. Mol. Biol. Cell. 2009;20:1949–1959. doi: 10.1091/mbc.E08-06-0549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miao H, Burnett E, Kinch M, Simon E, Wang BC. Activation of EphA2 kinase suppresses integrin function and causes focal-adhesion-kinase dephosphorylation. Nature Cell Biology. 2000;2:62–69. doi: 10.1038/35000008. [DOI] [PubMed] [Google Scholar]
- 28.Nasreen N, Mohammed KA, Lai Y, Antony VB. Receptor EphA2 activation with ephrinA1 suppresses growth of malignant mesothelioma (MM). Cancer Letters. 2007;258:215–222. doi: 10.1016/j.canlet.2007.09.005. [DOI] [PubMed] [Google Scholar]
- 29.Barquilla A, Pasquale EB. Eph receptors and ephrins: therapeutic opportunities. Annu. Rev. Pharmacol. Toxicol. 2015;55:465–487. doi: 10.1146/annurev-pharmtox-011112-140226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miao H, Li DQ, Mukherjee A, Guo H, Petty A, Cutter J, Basilion JP, Sedor J, Wu J, Danielpour D, Sloan AE, Cohen ML, Wang BC. EphA2 Mediates Ligand-Dependent Inhibition and Ligand-Independent Promotion of Cell Migration and Invasion via a Reciprocal Regulatory Loop with Akt. Cancer Cell. 2009;16:9–20. doi: 10.1016/j.ccr.2009.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miao H, Gale NW, Guo H, Qian J, Petty A, Kaspar J, Murphy AJ, Valenzuela DM, Yancopoulos G, Hambardzumyan D, Lathia JD, Rich JN, Lee J, Wang B. EphA2 promotes infiltrative invasion of glioma stem cells in vivo through cross-talk with Akt and regulates stem cell properties. Oncogene. 2015;34:558–567. doi: 10.1038/onc.2013.590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paraiso KH, Thakur MD, Fang B, Koomen JM, Fedorenko IV, John JK, Tsao H, Flaherty KT, Sondak VK, Messina JL, Pasquale EB, Villagra A, Rao UN, Kirkwood JM, Meier F, Sloot S, Gibney GT, STUART D, Tawbi H, Smalley KS. Ligand-Independent EPHA2 Signaling Drives the Adoption of a Targeted Therapy-Mediated Metastatic Melanoma Phenotype. Cancer Discov. 2015;5:264–273. doi: 10.1158/2159-8290.CD-14-0293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Binda E, Visioli A, Giani F, Lamorte G, Copetti M, Pitter KL, Huse JT, Cajola L, Zanetti N, DiMeco F, De Filippis L, Mangiola A, Maira G, Anile C, De Bonis P, Reynolds BA, Pasquale EB, Vescovi AL. The EphA2 Receptor Drives Self- Renewal and Tumorigenicity in Stem-like Tumor-Propagating Cells from Human Glioblastomas. Cancer Cell. 2012;22:765–780. doi: 10.1016/j.ccr.2012.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ireton RC, Chen J. EphA2 receptor tyrosine kinase as a promising target for cancer therapeutics. Current Cancer Drug Targets. 2005;5:149–157. doi: 10.2174/1568009053765780. [DOI] [PubMed] [Google Scholar]
- 35.Tandon M, Vemula SV, Mittal SK. Emerging strategies for EphA2 receptor targeting for cancer therapeutics. Expert Opinion on Therapeutic Targets. 2011;15:31–51. doi: 10.1517/14728222.2011.538682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rong BX, Cai XG, Yang SY, Li W, Ming ZJ. EphA2-Dependent Molecular Targeting Therapy for Malignant Tumors. Current Cancer Drug Targets. 2011;11:1082–1097. doi: 10.2174/156800911798073050. [DOI] [PubMed] [Google Scholar]
- 37.Wykosky J, Debinski W. The EphA2 Receptor and EphrinA1 Ligand in Solid Tumors: Function and Therapeutic Targeting. Molecular Cancer Research. 2008;6:1795–1806. doi: 10.1158/1541-7786.MCR-08-0244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Macrae M, Neve RM, Rodriguez-Viciana P, Haqq C, Yeh J, Chen CR, Gray JW, McCormick F. A conditional feedback loop regulates Ras activity through EphA2. Cancer Cell. 2005;8:111–118. doi: 10.1016/j.ccr.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 39.Singh DR, Ahmed F, King C, Gupta N, Salotto M, Pasquale EB, Hristova K. EphA2 Receptor Unliganded Dimers Suppress EphA2 Pro-tumorigenic Signaling. J. Biol. Chem. 2015;290:27271–27279. doi: 10.1074/jbc.M115.676866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koolpe M, Dail M, Pasquale EB. An ephrin mimetic peptide that selectively targets the EphA2 receptor. J. Biol. Chem. 2002;277:46974–46979. doi: 10.1074/jbc.M208495200. [DOI] [PubMed] [Google Scholar]
- 41.Riedl SJ, Pasquale EB. Targeting the Eph System with Peptides and Peptide Conjugates. Current Drug Targets. 2015;16:1031–1047. doi: 10.2174/1389450116666150727115934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mitra S, Duggineni S, Koolpe M, Zhu XJ, Huang ZW, Pasquale EB. Structure-Activity Relationship Analysis of Peptides Targeting the EphA2 Receptor. Biochemistry. 2010;49:6687–6695. doi: 10.1021/bi1006223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Duggineni S, Mitra S, Lamberto I, Han XF, Xu Y, An J, Pasquale EB, Huang ZW. Design and Synthesis of Potent Bivalent Peptide Agonists Targeting the EphA2 Receptor. Acs Medicinal Chemistry Letters. 2013;4:344–348. doi: 10.1021/ml3004523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang S, Placzek WJ, Stebbins JL, Mitra S, Noberini R, Koolpe M, Zhang ZM, Dahl R, Pasquale EB, Pellecchia M. Novel Targeted System To Deliver Chemotherapeutic Drugs to EphA2-Expressing Cancer Cells. J. Med. Chem. 2012;55:2427–2436. doi: 10.1021/jm201743s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barile E, Wang S, Das SK, Noberini R, Dahl R, Stebbins JL, Pasquale EB, Fisher PB, Pellecchia M. Design, Synthesis and Bioevaluation of an EphA2 Receptor-Based Targeted Delivery System. Chemmedchem. 2014;9:1403–1412. doi: 10.1002/cmdc.201400067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang NY, Fernandez C, Richter M, Xiao Z, Valencia F, Tice DA, Pasquale EB. Crosstalk of the EphA2 receptor with a serine/threonine phosphatase suppresses the Akt-mTORC1 pathway in cancer cells. Cell. Signal. 2011;23:201–212. doi: 10.1016/j.cellsig.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sinha B, Koster D, Ruez R, Gonnord P, Bastiani M, Abankwa D, Stan RV, Butler-Browne G, Vedie B, Johannes L, Morone N, Parton RG, Raposo G, Sens P, Lamaze C, Nassoy P. Cells respond to mechanical stress by rapid disassembly of caveolae. Cell. 2011;144:402–413. doi: 10.1016/j.cell.2010.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Raicu V, Stoneman MR, Fung R, Melnichuk M, Jansma DB, Pisterzi LF, Rath S, Fox M, Wells JW, Saldin DK. Determination of supramolecular structure and spatial distribution of protein complexes in living cells. Nature Photonics. 2009;3:107–113. [Google Scholar]
- 49.Biener G, Stoneman MR, Acbas G, Holz JD, Orlova M, Komarova L, Kuchin S, Raicu V. Development and Experimental Testing of an Optical Micro-Spectroscopic Technique Incorporating True Line-Scan Excitation. International Journal of Molecular Sciences. 2014;15:261–276. doi: 10.3390/ijms15010261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen LR, Novicky L, Merzlyakov M, Hristov T, Hristova K. Measuring the Energetics of Membrane Protein Dimerization in Mammalian Membranes. J. Am. Chem. Soc. 2010;132:3628–3635. doi: 10.1021/ja910692u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.King C, Stoneman M, Raicu V, Hristova K. Fully quantified spectral imaging reveals in vivo membrane protein interactions. Integr. Biol. (Camb. ) 2016;8:216–229. doi: 10.1039/c5ib00202h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sarabipour S, Del Piccolo N, Hristova K. Characterization of Membrane Protein Interactions in Plasma Membrane Derived Vesicles with Quantitative Imaging Forster Resonance Energy Transfer. Acc. Chem. Res. 2015;48:2262–2269. doi: 10.1021/acs.accounts.5b00238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Adair BD, Engelman DM. Glycophorin a helical transmembrane domains dimerize in phospholipid bilayers - a resonance energy transfer study. Biochemistry. 1994;33:5539–5544. doi: 10.1021/bi00184a024. [DOI] [PubMed] [Google Scholar]
- 54.Li M, Reddy LG, Bennett R, Silva ND, Jr., Jones LR, Thomas DD. A fluorescence energy transfer method for analyzing protein oligomeric structure: Application to phospholamban. Biophys. J. 1999;76:2587–2599. doi: 10.1016/S0006-3495(99)77411-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schick S, Chen LR, Li E, Lin J, Koper I, Hristova K. Assembly of the M2 Tetramer Is Strongly Modulated by Lipid Chain Length. Biophys. J. 2010;99:1810–1817. doi: 10.1016/j.bpj.2010.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sharonov GV, Bocharov EV, Kolosov PM, Astapova MV, Arseniev AS, Feofanov AV. Point Mutations in Dimerization Motifs of the Transmembrane Domain Stabilize Active or Inactive State of the EphA2 Receptor Tyrosine Kinase. J. Biol. Chem. 2014;289:14955–14964. doi: 10.1074/jbc.M114.558783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Noberini R, Lamberto I, Pasquale EB. Targeting Eph receptors with peptides and small molecules: Progress and challenges. Seminars in Cell & Developmental Biology. 2012;23:51–57. doi: 10.1016/j.semcdb.2011.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lin CC, Melo FA, Ghosh R, Suen KM, Stagg LJ, Kirkpatrick J, Arold ST, Ahmed Z, Ladbury JE. Inhibition of Basal FGF Receptor Signaling by Dimeric Grb2. Cell. 2012;149:1514–1524. doi: 10.1016/j.cell.2012.04.033. [DOI] [PubMed] [Google Scholar]
- 59.Ahmed Z, Lin CC, Suen KM, Melo FA, Levitt JA, Suhling K, Ladbury JE. Grb2 controls phosphorylation of FGFR2 by inhibiting receptor kinase and Shp2 phosphatase activity. J. Cell Biol. 2013;200:493–504. doi: 10.1083/jcb.201204106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chesi M, Brents LA, Fly SA, Bais C, Robbiani DF, Mesri E, Kuehl WM, Bergsagel PL. Activated fibroblast growth factor receptor 3 is an oncogene that contributes to tumor progression in multiple myeloma. Blood. 2001;97:729–736. doi: 10.1182/blood.v97.3.729. [DOI] [PubMed] [Google Scholar]
- 61.Chesi M, Nardini E, Brents LA, Schrock E, Ried T, Kuehl WM, Bergsagel PL. Frequent translocation t(4;14)(p16.3;q32.3) in multiple myeloma is associated with increased expression and activating mutations of fibroblast growth factor receptor 3. Nat. Genet. 1997;16:260–264. doi: 10.1038/ng0797-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Seo AN, Jin Y, Lee HJ, Sun PL, Kim H, Jheon S, Kim K, Lee CT, Chung JH. FGFR1 amplification is associated with poor prognosis and smoking in non-small-cell lung cancer. Virchows Archiv. 2014;465:547–558. doi: 10.1007/s00428-014-1634-2. [DOI] [PubMed] [Google Scholar]
- 63.Grandis JR, Sok JC. Signaling through the epidermal growth factor receptor during the development of malignancy. Pharmacology & Therapeutics. 2004;102:37–46. doi: 10.1016/j.pharmthera.2004.01.002. [DOI] [PubMed] [Google Scholar]
- 64.Tripp SR, Willmore-Payne C, Layfield LJ. Relationship between EGFR overexpression and gene amplification status in central nervous system gliomas. Analytical and Quantitative Cytology and Histology. 2005;27:71–78. [PubMed] [Google Scholar]