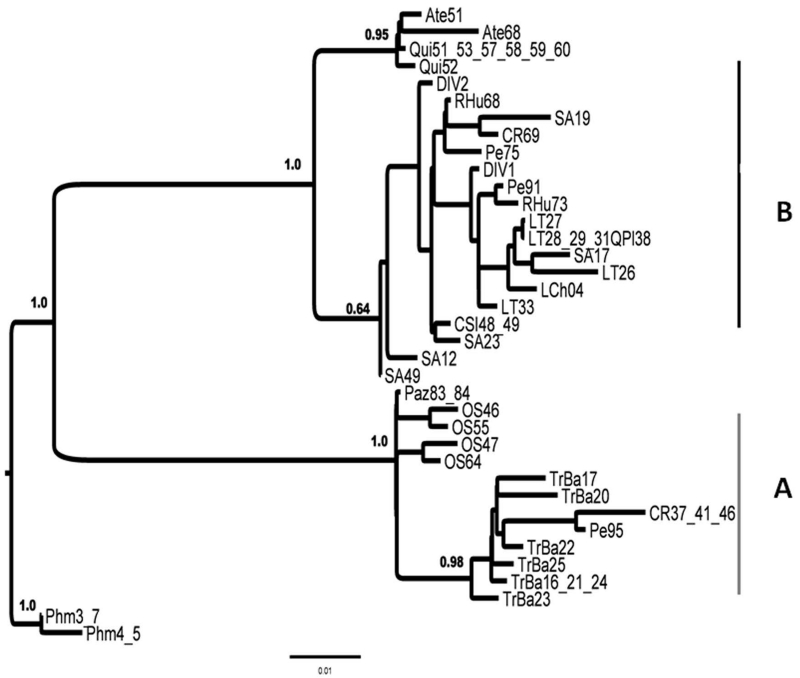
Figure 2.
Phyllotis darwini intraspecific phylogeny based on Bayes Markov Chain Monte Carlo method (BMCMC). The phylogeny was obtained for the Hypervariable Domain II (HV2) from the mitochondrial control region sequence data, whereas for BMCMC represents a consensus tree from the n = 9950 trees from the converged Markov chain. Posterior probability values over 0.5 are represented on each node.