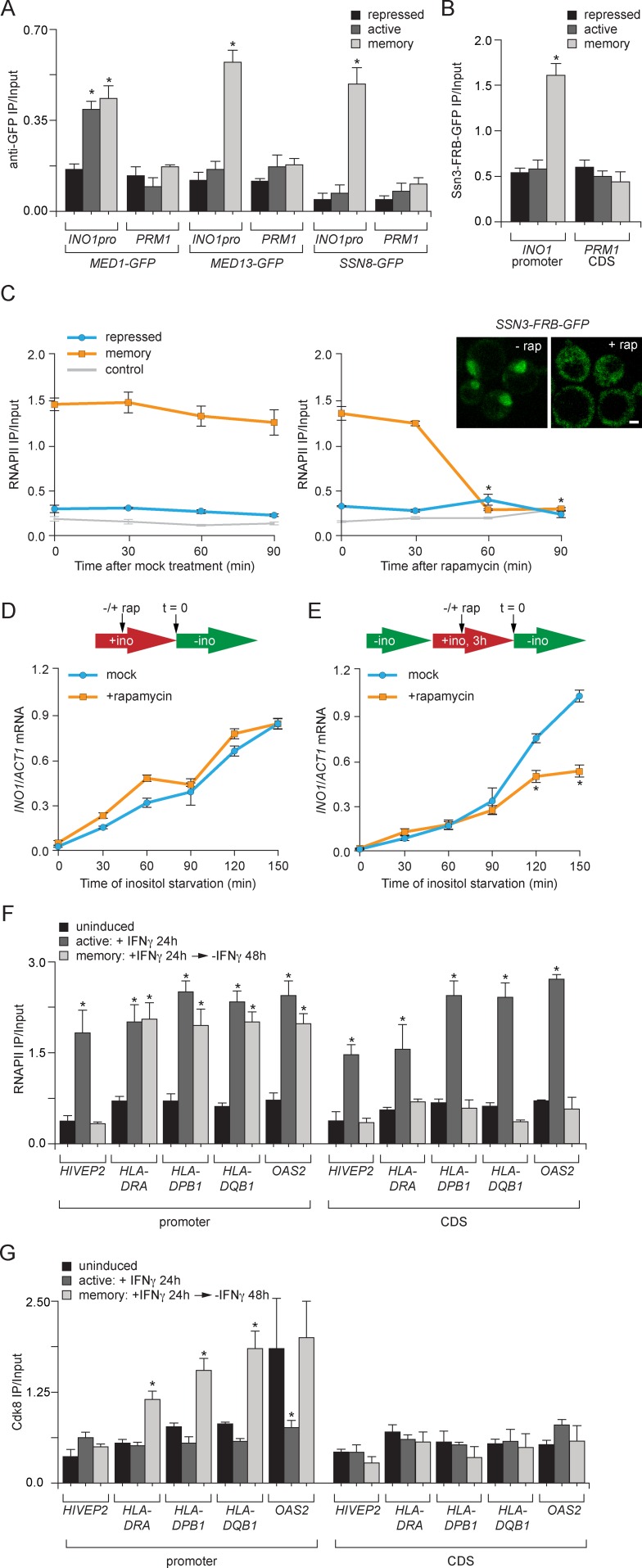
Figure 6. Transcriptional memory leads to Ssn3/Cdk8-dependent poised preinitiation complex.
(A) ChIP against Med1-GFP, Med13-GFP or Ssn8-GFP from cells grown under repressing, activating or memory conditions. (B) ChIP against Ssn3-FRB-GFP from cells grown in repressing, activating or memory conditions. (A and B) *p<0.05, compared with the repressing condition (Student’s t-test). (C) ChIP against RNAPII from strains expressing Ssn3-FRB-GFP grown under either repressing or memory conditions, before or after treatment 1 µg/ml of rapamycin using primers to amplify the INO1 promoter (−348 to −260) or the PRM1 CDS. Inset: confocal micrographs of Ssn3-FRB-GFP expressing cells before or after treatment with 1 mg/ml of rapamycin for 30 min. *p<0.05, compared with t = 0 (Student’s t-test). (D and E) INO1 activation (D) or reactivation (E) in Ssn3-FRB-GFP cells. For activation at time = 0, cells were shifted from medium containing 100 μM inositol (repressing conditions; red arrow in schematic) to medium without inositol (activating conditions; green arrow in schematic). For reactivation, cells were shifted from activating medium to repressing medium containing 100 μM inositol for 3 hr. Cells were treated ±1 µg/ml rapamycin for 45 min before transferring to activating conditions. Cells were harvested at the indicated time points, and INO1 mRNA levels were quantified relative to ACT1 mRNA levels by RT-qPCR. The averages of three biological replicates are shown ± standard error of the mean. *p<0.05, compared with the same time point in the mock-treated culture (Student’s t-test). (F and G) ChIP against RNAPII (F) or Cdk8 (G) from HeLa cells before, during (24 hr) or 48 hr after treatment with 50 ng/mL Interferon-γ. Recovery of the indicated promoters or coding sequences (CDS) of genes that exhibit transcriptional memory (HLA-DRA, HLA-DPB1, HLA-DQB1 and OAS2) and a gene that does not (HIVEP2) was quantified relative to input by qPCR. *p<0.05, compared with the uninducing condition (Student’s t-test). (A-F) Averages of three biological replicates ± standard error of the mean.