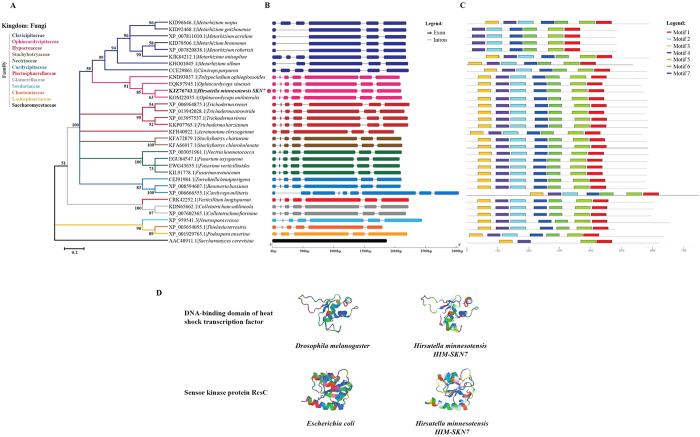
Figure 1. Phylogenetic analysis, gene structure and motifs comparison.
(A) Phylogenetic analysis of the different orthologs of HIM-SKN7 (KJZ76743.1|Hirsutella minnesotensis SKN7). The protein sequences were aligned with ClustalW, and a Maximum likelihood (ML) tree was generated using a WAG model. (B) Genomic and coding sequences were used for gene structure analysis to identify the number of introns and exons. (C) The motifs analysis comparison showed various conserved regions in the protein sequences of HIM-SKN7 and its different orthologs. (D) Prediction of the tertiary structure of the conserved domain in HIM-SKN7 and comparison with that of Drosophila melanogaster and Escherichia coli.