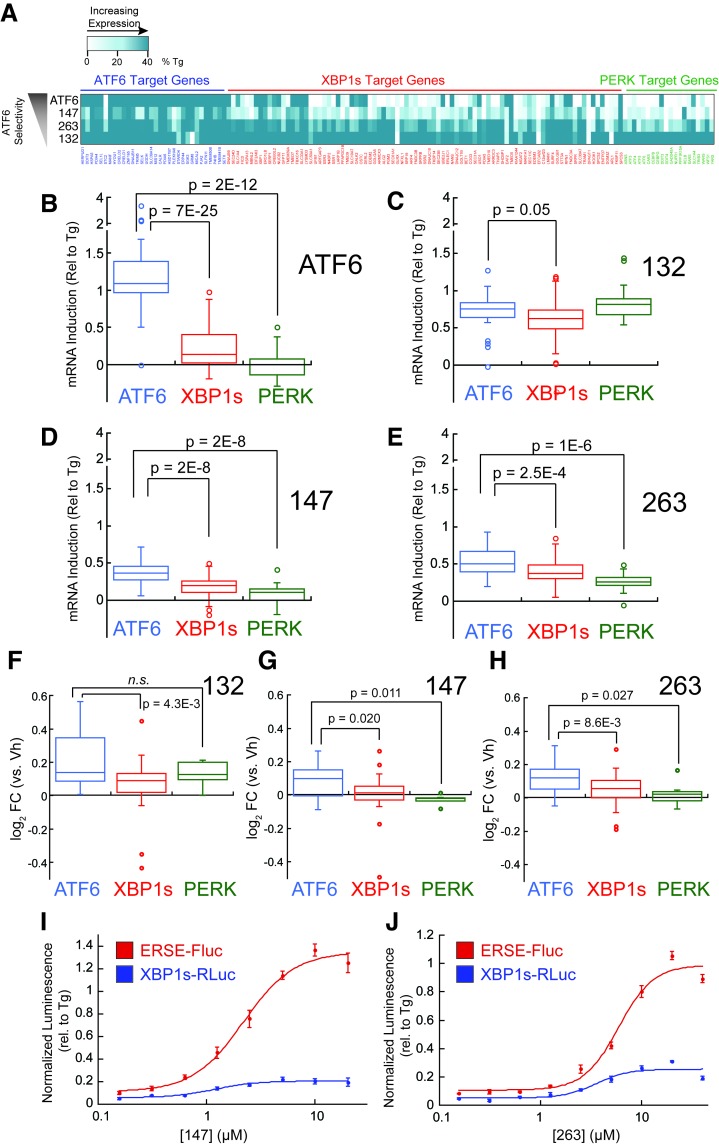
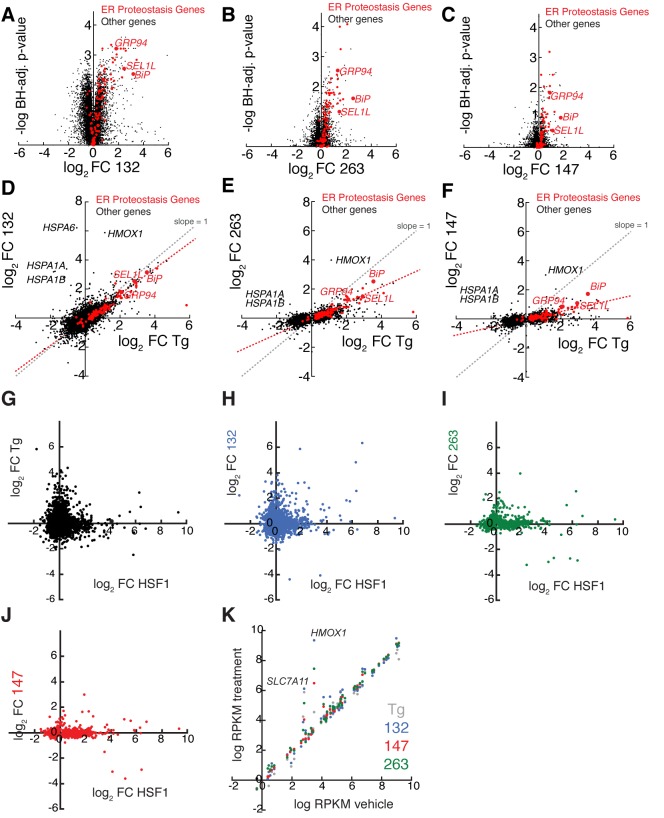
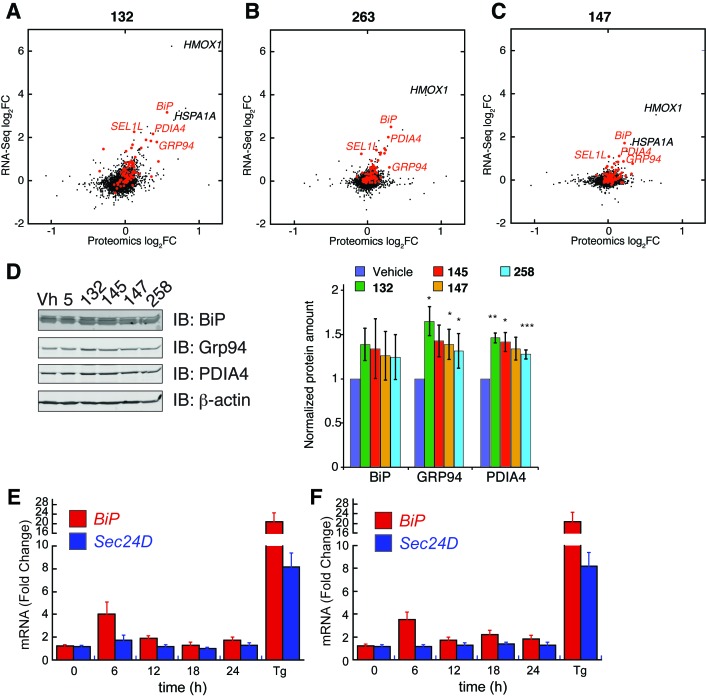
Figure 3. Small molecule ER proteostasis regulators preferentially activate the ATF6 transcriptional program.
(A) Heat map showing the upregulation of ATF6 (blue), XBP1s (red) or PERK (green) target genes in the mRNA-Seq analysis of HEK293T-Rex cells treated with Tg (1 µM), 132 (10 µM), 147 (10 µM) or 263 (10 µM). Genes used in this analysis were selected from published reports indicating their selectivity for ATF6, XBP1s or PERK (Lu et al., 2004; Shoulders et al., 2013). The induction of each gene was normalized to the respective induction observed with the global UPR activator Tg and reported as % Tg induction. The expression of these target genes in the mRNA-Seq analysis of TMP-dependent DHFR-ATF6 activation in HEK293DAX cells is also shown. The data used to prepare this heat map is included in Figure 3—source data 1. (B–E) Box plots showing the relative activation of ATF6, XBP1s, and PERK genesets in HEK293DAX cells following TMP-dependent DHFR-ATF6 activation (B), or HEK293T-Rex cells treated with 132 (C), 147 (D) or 263 (E) from the data shown in Figure 3A. Differences in activation of the ATF6 geneset relative to the XBP1s or PERK genesets were confirmed by one-way ANOVA and the p-values of unpaired t-tests are shown. (F–H) Box plots comparing the average fold change at the protein levels of ATF6, XBP1s and PERK target sets in HEK293T-Rex cells treated with 10 µM 132 (F), 147 (G), or 263 (H) for 16 hr as observed in the proteomics analysis. The protein sets used here are the same as the genesets for the analysis of the RNA-Seq data (A–E). Differences in activation of the ATF6 target protein set relative to the XBP1s or PERK target protein sets were confirmed by one-way ANOVA and the p-values of unpaired t-tests are shown. The data used to prepare these graphs can be found in Figure 3—source data 1. (I–J) Plot showing the relative activation of the ERSE-FLuc (blue) and XBP1-RLuc (red) reporters in HEK293T-Rex cells treated with the indicated concentration of 147 (I) or 263 (J) for 15 hr. Error bars represent standard error for n = 3 replicates.
DOI: http://dx.doi.org/10.7554/eLife.15550.010