Table 1.
Prioritized ER proteostasis regulators.
| Name | Structure | MW | Polar Surface Area | AlogP | Structural moieties (Figure 1—figure supplement 1B–C) |
% ERSE-FLuc activation* | % XBP1-RLuc activation* | Promiscuity** |
|---|---|---|---|---|---|---|---|---|
|
5 |
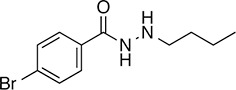 |
271.2 |
41.13 |
2.62 | - | 60.9 ± 6.0 | 9.1 ± 0.7 | 1 out of 31 |
| 132 | 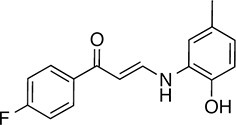 |
283.3 | 58.56 | 3.27 | F | 140.1 ± 17.4 | 10.3 ± 7.9 | 2 out of 32 |
| 145 | 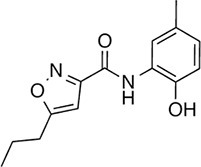 |
260.3 | 75.36 | 3.01 | F, H | 83.4 ± 6.3 | 24.5 ± 4.7 | 1 out of 31 |
| 147 | 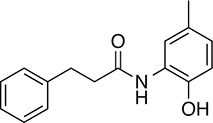 |
255.3 | 49.33 | 3.35 | B, F | 63.2 ± 0.6 | 17.5 ± 1.9 | 1 out of 32 |
| 148 | 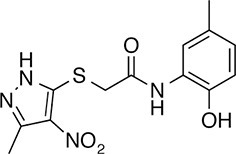 |
322.3 | 149.12 | 2.00 | B, F | 52.5 ± 14.3 | 16.6 ± 2.4 | 3 out of 31 |
| 238 | 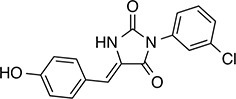 |
314.7 | 69.64 | 2.94 | B, L | 68.2 ± 5.7 | 61.1 ± 0.1.5 | 2 out of 31 |
| 258 | 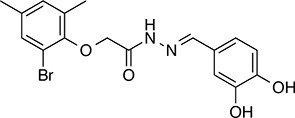 |
393.2 | 91.15 | 3.76 | C, D | 28.6 ± 6.1 | 32.6 ± 1.0 | 1 out of 31 |
| 263 | 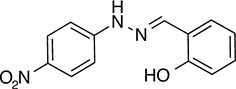 |
257.2 | 90.44 | 3.10 | C | 20.7 ± 2.3 | 2.4 ± 1.0 | 1 out of 31 |
*% Tg activation from triplicate confirmation screen at 6.8 μM.
** Number of screening assays in which the compound was found active.
