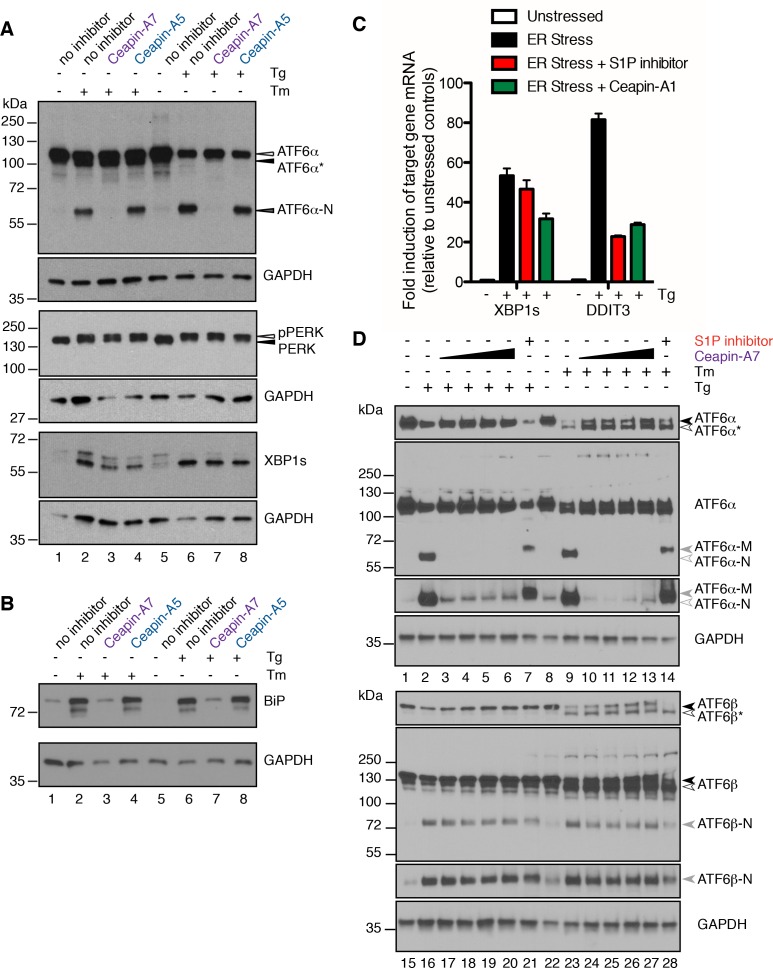
Figure 5. Ceapins are selective inhibitors of the ATF6α branch of the UPR.
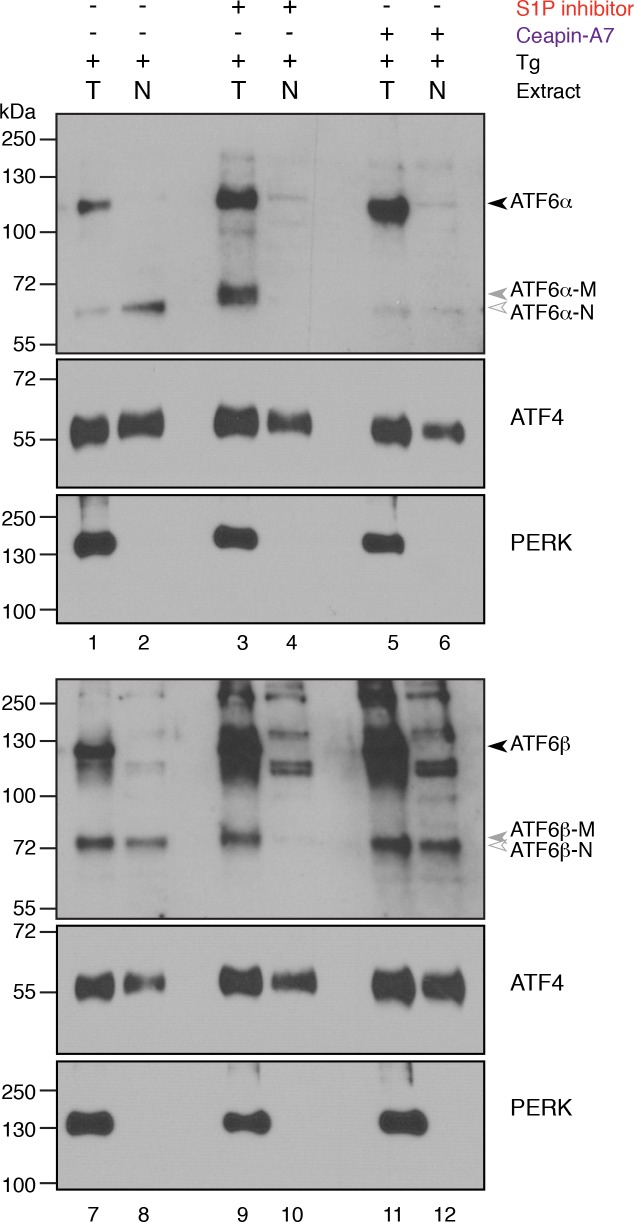
(A) U2-OS cells were treated without (DMSO) or with ER stressor (100 nM Tg or 2.5 μg/mL Tm) in the absence or presence of Ceapin analogs (6 μM each) for two hours. One hour prior to lysis, proteasomal inhibitor (10 μM MG132) was added to prevent the degradation of the cleaved ATF6α-N fragment. Cells were harvested and analyzed by Western Blot for ATF6α, PERK, XBP1 and GAPDH (loading control). Arrowheads denote the positions of full-length (ATF6α), unglycosylated full-length (ATF6α*) and cleaved (ATF6α-N) variants of ATF6α and also PERK and phospho-PERK. Data shown is representative of three independent experiments. (B) U2-OS cells were treated without (DMSO) or with ER stressor (100 nM Tg or 2.5 μg/mL Tm) in the absence or presence of Ceapin analogs (6 μM each). After eight hours, cells were harvested and analyzed by Western Blot for BiP and GAPDH (loading control). (C) U2-OS cells were treated without (DMSO) or with ER stressor (100 nM Tg, black) in the absence or presence of ten-fold the IC50 of either S1P inhibitor (Pf-429242, 3.2 μM, red) or Ceapin-A1 (35.7 μM, green). Four hours later cells were harvested and mRNA extracted. mRNA levels for XBP1s or DDIT3 were normalized to GAPDH for each well and then compared to unstressed controls. Data plotted are from duplicate qPCR reactions from duplicate wells, error bars are standard deviation. (D) U2-OS cells were treated without (DMSO) or with ER stressor (100 nM Tg or 2.5 μg/mL Tm) in the absence or presence of increasing concentration of Ceapin-A7 (0.6, 1.89, 6, 18.9 μM) or S1P inhibitor (5 μM Pf-429242) for four and a half hours. One hour prior to lysis, proteasomal inhibitor (10 μM MG132) was added to prevent the degradation of the cleaved ATF6α-N and ATF6β-N fragments. Cells were harvested and analyzed by Western Blot for ATF6α, ATF6β and GAPDH (loading control). Arrowheads denote the positions of full-length (ATF6α, ATF6β), unglycosylated full-length (ATF6α*, ATF6β*), cleaved membrane bound (ATF6α-M) and cleaved (ATF6α-N, ATF6β-N) variants of ATF6α and ATF6β. Data shown is representative of three independent experiments.
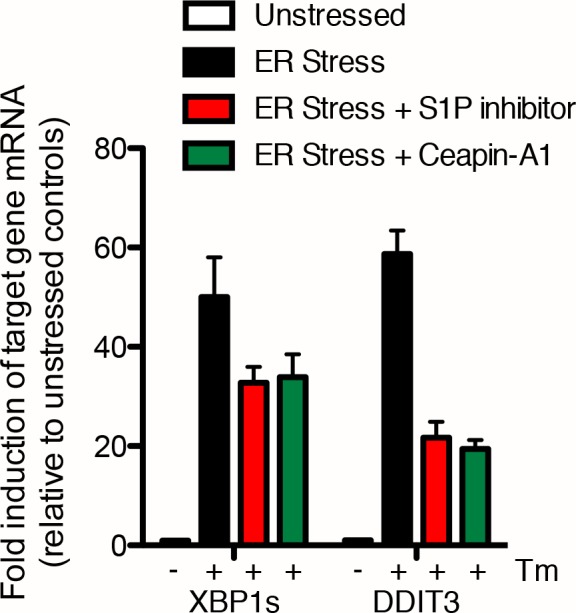
Figure 5—figure supplement 1. Induction of XBP1s and DDIT3 mRNA is only partially inhibited by either the S1P inhibitor or Ceapin-A1 U2-OS cells were treated without (DMSO) or with ER stressor (2.0 μg/mL Tm, black) in the absence or presence of ten-fold the IC50 of either S1P inhibitor (Pf-429242, 3.2 μM, red) or Ceapin-A1 (35.7 μM, green).