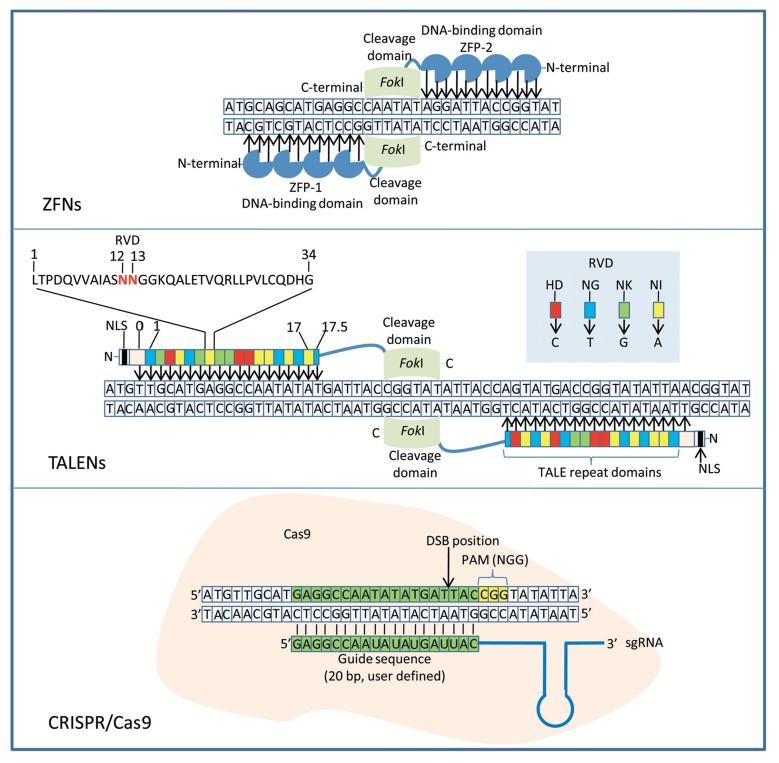
Fig. (1).
Three types of engineered nucleases used for genome engineering in plants. Zinc finger nucleases (ZFNs) recognise and bind to target sites on DNA. Binding of both ZFN-1 and ZFN-2 leads to dimerization and activation of the FokI endonuclease catalytic domains and subsequent cleavage of target site. Transcription activator-like effector nucleases (TALENs) are made up of repeat domains each with its specific nucleotide binding specificity depending on the Repeat di-variable residues (RVD) present on position 12 and 13 on each repeat. The ‘0’ domain specifically binds thymine only. Binding of left and right TALENs result in FokI catalytic domain dimerization, which leads to cleavage resulting in a double-strand break on the DNA target site. The CRISPR/Cas9 system is guided by an engineered sgRNA, which is composed of the crRNA and the tracrRNA. The crRNA contains a customizable 20-nucleotide guide RNA at its 5’-end which binds to target sites on the DNA and cleavage by Cas9 occurs within the target site adjacent to the protospacer adjacent motive (PAM).