Abstract
Nucleic acids are among the most researched and applied biomolecules. Their diverse two- and three-dimensional structures in conjunction with their robust chemistry and ease of manipulation provide a rare opportunity for sensor applications. Moreover, their high biocompatibility has seen them being used in the construction of in vivo assays. Various nucleic acid-based devices have been extensively studied as either the principal element in discrete molecule-like sensors or as the main component in the fabrication of sensing devices. The use of aptamers in sensors - aptasensors, in particular, has led to improvements in sensitivity, selectivity, and multiplexing capacity for a wide verity of analytes like proteins, nucleic acids, as well as small biomolecules such as glucose and adenosine triphosphate (ATP). This article reviews the progress in the use of aptamers as the principal component in sensors for optical detection of ATP with an emphasis on sensing mechanism, performance, and applications with some discussion on challenges and perspectives.
Keywords: Adenosine triphosphate, nucleic acid, aptamers, aptasensors, fluorometry, colorimetry, chemiluminescence.
1. Introduction
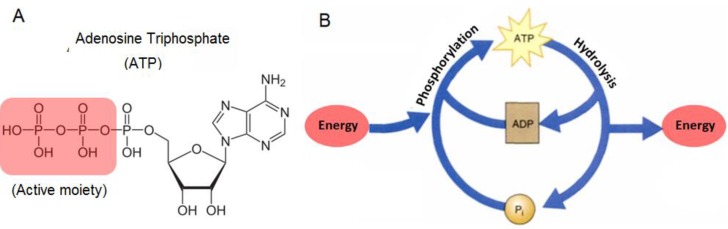
Adenosine triphosphate (ATP) is an important biological molecule in all living organisms. It is made up of three subunits, namely an adenine base, a 5-carbon ribose sugar, and a triphosphate (Figure 1A). The combination of the ribose sugar and the adenine forms the adenosine component of ATP while the triphosphate end forms the reactive site for ATP to take part in various cellular reactions. In living organisms, ATP is typically produced in a series of reactions in the presence of enzymes, mainly in the mitochondria and chloroplasts of living cells. The main function of ATP is as its usage as a major form of energy currency in living organisms. This unique property of ATP is attributed to the 'active region' or the reactive triphosphate end of ATP. Due to the low activation energy required to break the relatively weak phosphor-oxygen bond, the end phosphate group can be readily transferred from one molecule to another while simultaneously releasing useful energy for work. When a phosphate group is removed from ATP and transferred to a target molecule, the resulting products formed are adenosine diphosphate (ADP) and a phosphorylated target molecule.
Figure 1.
(A) Chemical structure of ATP and (B) the ATP-ADP cycle.
Since at any one point in time, the cellular system contains a pool of ATP, ADP, phosphorylated molecules, and energy molecules, the ATP-ADP cycle is continuously regenerated. This energy cycle shown in Figure 1B, plays a vital role in the regulation of cellular metabolism and biochemical processes in cellular physiology 1,2. ATP is also needed for building of large molecules like proteins from amino acids, active transport of ions across different parts of living organisms, and as electrical signals in the nervous system synaptic transmission. In addition to its use as energy currency in cells, the presence of ATP in cells has also been monitored as an indication of the cell's viability and the presence of cellular injuries. This is because ATP exists in all living cells and will quickly diminish once the cells die off. For example, the participation of ATP in critical cellular processes such as metabolism, active transport, and mechanical work of muscular cells, makes it necessary to accurately measure ATP levels in order to investigate the biochemical, structural, and functional indications of heart blockage 3. Not only is this important for human health, but also ATP has been frequently used to determine microbial activity in soil materials, the freshness of fish, and as a quality control of the functionality of blood prior to transfusion. In addition, recent studies have indicated that ATP is one of the most important chemical signaling agents, playing a central role in signal transduction 4-6 and one type of neurotransmitters that is related to the sense of taste 7,8.
Given the attention bestowed onto ATP in so many areas, it has garnered a wide interest in analytical chemistry, especially in the methods used to accurately and reproducibly determine its concentration. These included biosensors employing peptides 9, conjugated polymers 10, host-guest receptors 11, and ATP-dependent bioluminescence. Among them, the ATP-dependent bioluminescence method is the most studied and most sensitive. In this ATP-dependent reaction, ATP is required as the energy source for the bioluminescence to occur. The reaction is sensitive and highly specific to ATP. During this process, the enzyme luciferase, magnesium ion, and molecular oxygen are required. Light is emitted as a result of this reaction with its intensity directly correlates to the concentration of ATP. This method has shown its usefulness in the detection of ATP in most biological samples but not without limitations. Despite the ability of achieving a detection limit as low as 10-15g 12, the enzyme luciferase has shown to be unstable and hence unlikely to be used for the detection of ATP under conditions that are drastically different from physiological conditions. In addition, some biological substances such as plasma and ATPase degrade ATP readily. Also, there may be the possibility of overestimation of ATP concentration with possible contributions from red blood cells and platelets present in biological samples.
With these limitations, there have been continuous efforts to develop other ATP detection methods hoping to overcome some of the limitations. In the 1990s, specific single-stranded nucleic acid sequences for selectively interacting with certain targets had first been produced by systematic evolution of ligands by exponential enrichment (SELEX) 13 and repeated rounds of in vitro selection 14. These sequences are now known as aptamers, which can fold into specific three-dimensional structures and selectively bind to their targets. As for ATP, the first RNA and DNA ATP aptamers were reported by Szostak and colleagues 15,16. It was shown that both the sugar and the base are involved in the interaction with the aptamers and the binding of ATP accompanies with some conformational changes of the aptamers. Further structural study indicated that the DNA and RNA ATP aptamers have distinctly different tertiary structures and binding stoichiometries although they share similar structures and recognition sites in their binding pockets but also with considerable affinity towards other adenosine monophosphates 17. Basically, there are three types of ATP aptamers, namely, simple single-stranded nucleotide aptamer, split aptamer, and structural switching aptamers with the possibility of incorporating additional functionality like catalytic ribozyme and DNAzyme. In addition, several reports have shown that some ATP aptamers are allosterically catalytic. For example, Breaker's group assembled several ribozymes and DNAzymes by modular rational design - coupling catalytic domains and receptor domains through structural bridges 18-24. One of the constructed DNAzymes is an ATP-dependent DNAzyme with kinase activity, operating 109-fold faster than uncatalyzed rate of ATP hydrolysis 24. Other examples of allosteric ribozymes and DNAzymes can be found in the works of Sen et al. 25 and Ellington et al. 26,27. It has been shown that the allosteric ATP-dependent ribozymes and DNAzymes respond either positively 24-26 or negatively 22 to the presence of ATP (the allosteric effector). The allosteric regulation of the catalytic activity of aptamers (ribozymes and DNAzymes) by ATP operates by mechanisms that are fundamentally different from that of inhibitors. ATP binds to a site located apart from the active site and its influence on the catalytic activity of the aptamer is primarily originated from the conformational change of the aptamers 18,19.
Leveraging on their high affinity and catalytic activity, aptamers were quickly employed in the construction of biosensors - aptasensors. Considering the low stability and high cost of RNA aptamers, most of aptasensors engage DNA aptamers. Very often, conformational changes of aptamers upon target binding, which alter the surrounding environment, produce measurable signals. Aptamers have attractive advantages such as very small molecular mass, high stability, ease of synthesis and modification with functional materials such as fluorophores, and diversity of their targets. A wide variety of approaches have been developed for the detection of biomolecules 28,29.
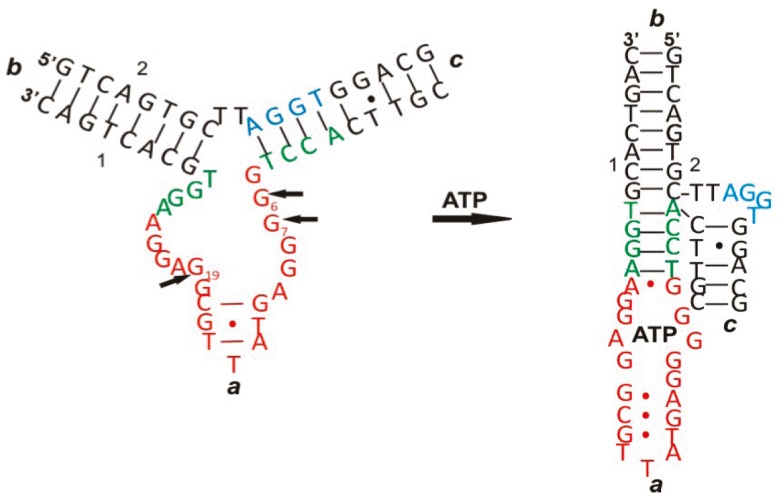
ATP is one of the important biomolecules of interest in the development of aptamer-based biosensors - ATP aptasensors 30. Like other aptamers, binding of ATP to ATP aptamers involves a series of intramolecular and intermolecular interactions among nucleotides and between nucleotides and ATP such as aromatic stacking and hydrogen bonding, thus leading to both conformational and structural changes. An example is given in Figure 2 31. As seen in Figure 2, there is a drastic change in its two-dimensional and three-dimensional conformation upon interacting with ATP. Therefore, many of the ATP aptasensors exploit this change to devise their signal generation mechanism.
Figure 2.
The conformational and structural change of an ATP aptamer upon binding to ATP. (Reproduced with permission from 31. ©2011 American Chemical Society)
This article intends to provide a broad coverage of the optical ATP aptasensors and their possible applications. The sensing principles of the optical ATP aptasensors, such as fluorescent, colorimetric, and chemiluminescent ATP aptasensors, will be discussed in detail. The progress in the research and development of the optical ATP aptasensors are summarized and the advantages and disadvantages of each type of the optical aptasensors as well as possible future work will also be discussed. It is hoped that this article will provide a comprehensive overview of the current status of the optical ATP aptasensors research and open new perspectives towards the research and applications of the optical ATP aptasensors with much improved analytical performance.
2. Fluorescent aptasensors
Fluorescence involves the absorption of light by a fluorophore (fluorescent molecule or quantum dot), which excites the fluorophore to promote electrons from ground state to excited states. The electrons often rapidly relax to the lowest energy level of the excited states and then emit light of longer wavelength when they relax from the lowest energy level of the excited states to the ground state. Techniques involving fluorescence detection have the advantage of good sensitivity. However, being essentially nucleic acids, aptamers are not inherently fluorescent, it is necessary to modify the aptamers with fluorophores or build a fluorescence generation mechanism so that they can have fluorescence property. Fluorescent ATP aptasensors can be broadly classified into labeled and non-labeled aptasensors. They will be discussed in the following sections. Generally, the performance of the labeled aptasensors is better than their non-labeled counterparts. On the other hand, major downsides of the labeled aptasensors are their high cost and sometime time-consuming multistep protocols.
2.1. Non-amplified fluorescent ATP aptasensors
The first group of non-amplified fluorescent ATP aptasensors is based on non-labeled signal generation strategy. The non-labeled fluorescent ATP aptasensors involve the use of aptamers in conjunction with fluorescent intercalating dyes 32-34, conjugated polymers 35,36, nanoparticles 37, and nanoclusters 38,39. The non-labeled aptasensors have gained increasing popularity since they are usually cheaper and simpler than the labeled aptasensors. Moreover, the non-labeled aptasensors do not have the risk of binding site alteration when fluorophores are covalently bonded to aptamers.
Intercalating dyes, for example [Ru(phen)2(dppz)]2+ 40 and ethidium bromide 41 have been investigated for binding to aptamers, including in the application of ATP detection. In aqueous solutions, [Ru(phen)2(dppz)]2+ has no fluorescence. However, fluorescence is observable when [Ru(phen)2(dppz)]2+ intercalates with aptamers. The binding of ATP to its aptamer induces an alteration in conformation and hence alters the fluorescence intensity of the intercalated [Ru(phen)2(dppz)]2+ 42. Unfortunately, the magnitude of the fluorescence alteration is not significant enough to have a sensitive ATP aptasensor. To enhance fluorescence intensity, Wang et al. explored the use of metal-enhanced fluorescence (MEF) for ATP detection using PicoGreen as the intercalating dye. MEF serves to enhance the quantum yield of PicoGreen and provides photostability 43. Silver island films are used as MEF in this strategy. An aptamer is pre-hybridized with its complementary DNA strand and PicoGreen intercalates in the hybridized aptamer. Fluorescence is enhanced in proximity to the silver island films. The presence of ATP induces a conformational change during the formation of the ATP-aptamer complex and some of the intercalated PicoGreen is released 44. A similar PicoGreen release mechanism upon ATP binding was also engaged in a more recent work 45. Moreover, a cationic tetrahedralfluorene was also reported to increase the sensitivity and selectivity of ATP aptasensors using ethidium bromide as the intercalating dye. A 10-fold enhancement in sensitivity was obtained when tetrahedralfluorene serving as an energy donor to ethidium bromide 46.
Polymers with extended conjugation have the ability to rapidly transport charges through their conjugated backbones of repeating monomer units. This property deemed it attractive for use in biosensors, allowing enhanced detection sensitivity. Poly(2,5-bis(3-sulfonatopropoxy)-1,4-phenyl ethynylenealt-1,4-poly(phenylene ethynylene) (PPESO3) is such a conjugated polymer, which is water soluble and has fluorescent properties. In the work by Yang et al., PPESO3 was investigated for ATP detection 47. Cu2+ is used as quencher for this conjugated polymer by strong electrostatic attraction and electron transfer. ATP disrupts the electrostatic interaction between PPESO3 and Cu2+; hence the recovery of fluorescence is observed. This strategy was further demonstrated to be able to quantitatively measure the recovery efficiency in real human blood samples using a linear regression plot.
Apart from solution processes and surface-immobilization, as demonstrated by Brennan's lab, sol-gel-derived materials are a promising matrix for the entrapment of ATP aptamers and the development of fluorescent ATP aptasensors with improved analytical performance as compared to surface-immobilized ATP aptamers 48,49. Further investigation demonstrated that the analytical performance and stability of the aptasensors are primarily determined by the ability of the aptamers to remain fully hybridized to their signaling units upon entrapment 50.
The second group of non-amplified fluorescent ATP aptasensors leverage on the use of fluorophore-labeled aptamers. This fluorescence generation mechanisms can be classified into mono- and bichromophore approaches 51. The monochromophore approach refers to the covalent binding of a single fluorophore to an aptamer at a site where target binding induces a significant conformational change and this change results in a fluorescence intensity change. The bischromophore approach involves the attachment of two molecules, a fluorophore and a quencher, onto an aptamer. Fluorescence intensity changes when target binding to the aptamer alters the distance between the fluorophore and the quencher. This is also known as fluorescence resonance energy transfer (FRET), in which fluorescence quenching is observed when the fluorophore and quencher are in close proximity. The fluorophore and quencher could also be attached separately on an aptamer and its complementary strand. In the presence of the target, preferential binding of the target to the aptamer triggers the dehybridization between the two strands and hence FRET is terminated when the distance between the fluorophore and quencher widens.
Zeng et al. proposed an ATP aptasensor based on a molecular beacon probe 52. In the absence of ATP, fluorescence quenching occurs with close proximity between the fluorophore and quencher on the two ends of the molecular beacon probe. An oligonucleotide strand, partially complementary to both the molecular beacon probe and the aptamer, preferentially binds to the aptamer over the molecular beacon probe. However, in the presence of ATP, the aptamer instead binds to ATP and the oligonucleotide strand is released to bind to the molecular beacon probe. This induces a conformational change and the separation of the fluorophore and quencher. Consequently, fluorescence is observed and its intensity directly correlates to the concentration of ATP. This aptasensor was shown to have high selectivity for ATP compared to its close analogues, cytidine triphosphate (CTP), guanosine triphosphate (GTP), and uridine triphosphate (UTP). In another report, a double-receptor sandwich supramolecule was studied by Zhao et al. 53. A uranyl-salophen complex, which recognizes specifically the triphosphate moiety of ATP, is first immobilized onto silica gel nanoparticles. ATP is then binds to the immobilized complex. The binding of fluorophore-labeled ATP aptamer to ATP bound on the complex forms a sandwich supramolecule, granting the supramolecule fluorescence.
2.2. Amplified fluorescent ATP aptasensors
Although the amplification-free fluorescent ATP aptasensors offer simple and straightforward signal readouts, only moderate sensitivity could be achieved for the above-mentioned two groups of aptasensors. Hence, amplification strategies have to be explored to improve the sensitivity for the detection of trace amounts of ATP. Much effort has therefore been devoted to overcome these problems, and some of the improvements will be elaborated in the following section.
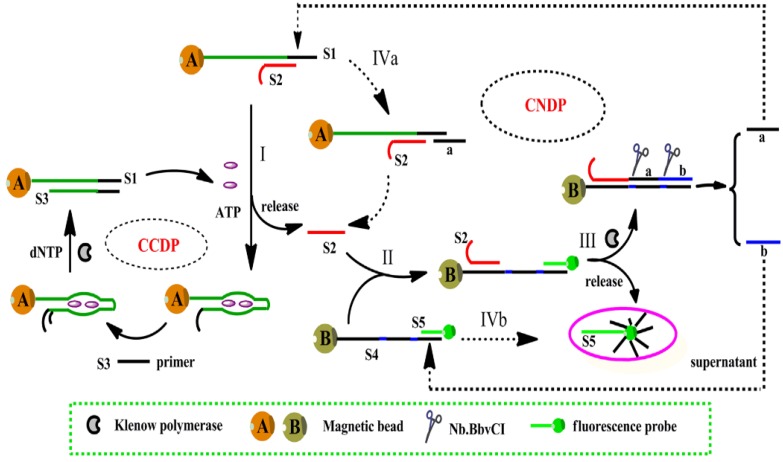
One of the strategies was proposed by Song et al., based on isothermal circular strand-displacement polymerization (Figure 3) 54. The process is made up of circular common target molecule-displacement polymerization (CCDP) and circular nucleic acid strand-displacement polymerization (CNDP). In this aptasensor, the complementary strand (S2) is released when ATP binds to a recognition probe (S1). Subsequently, CNDP is triggered by the released S2. In CNDP, a fluorescent probe S5 is displaced by the hybridization of S2 to DNA S4, fluorescence is hence generated. Scission of strand a and b occurs with the formation of a duplex between S2 and S4. Strand a then hybridizes to S1 and S2 is released during polymerization, which triggers the CNDP cycle over again. On the other hand, strand displacement process occurs to generate a fluorescent probe when strand b hybridizes to S4. CNDP occurs concurrently, releasing ATP from its aptamer when a DNA duplex between S1 and S3 forms in the presence of a primer S3. The continuous processes of both CNDP and CCDP thus generate a large amount of fluorescent probes in the presence a minute amount of ATP. This approach achieved a detection limit down to 0.26 nM. However, this technique is more complex and expensive, requiring more materials such as polymerase and primers.
Figure 3.
Schematic representation of ATP detection by isothermal circular strand-displacement polymerization. (Reproduced with permission from Reference 54. © 2014 Elsevier Ltd)
Another amplification strategy was developed by engaging rolling circle amplification (RCA) 55. As mention earlier, allosterically catalytic ATP aptamers can be constructed by modular rational design. The catalytic power of the aptamers can be utilized in the configuration of amplification strategies. As described by Cho and colleagues 55, an ATP-dependent allosteric DNAzyme is first immobilized on a glass slide. The DNAzyme is activated upon binding to ATP. Consequently, a padlock probe is ligated and RCA is initiated from 3' end of the DNAzyme. After RCA, Cy-3 labelled short oligonucleotides fully complementary to the repetitive sequences of the RCA product are introduced to generate a fluorescence signal. In a more recent report, reduced graphene oxide was coupled to RCA in the development of fluorescent ATP aptasensors 56. Upon binding to ATP, the ATP aptamer is released from the reduced graphene oxide due to a conformational change of the aptamer. The released aptamer initiates a round of RCA, thus producing a detection limit as low as 60 nM. In addition, the binding of ATP to an inactivated DNAzyme containing ATP aptamer 57 or an ATP aptamer partially hybridized with a DNAzyme 58 can trigger the release of the active DNAzyme and the released DNAzyme can be engaged in the configuration of amplified detection of ATP.
2.3. Nanomaterial-assisted fluorescent ATP aptasensors
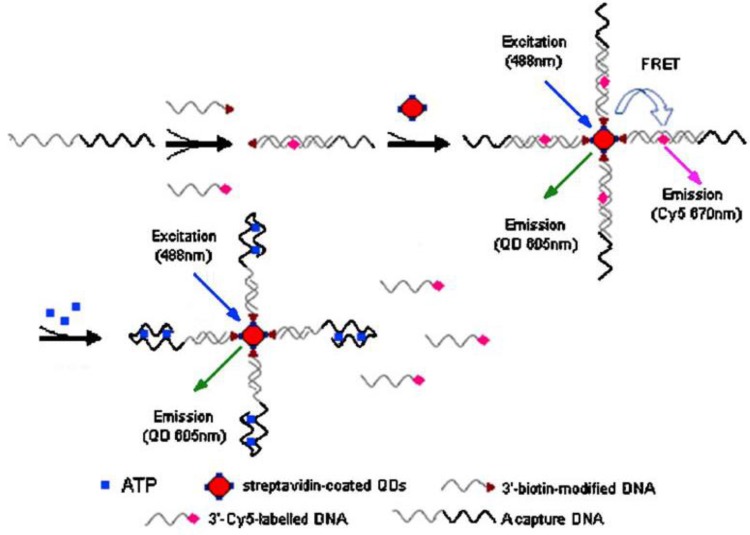
Because of extremely large surface-to-volume ratios, diverse surface morphologies and functionalities, high dispersibilities, and excellent size-dependent properties such as surface plasmon resonance and quantum confinement, nanomaterials have found interesting applications in the fluorescent ATP aptasensors, For example, silica nanoparticles have been widely used for the detection of nucleic acids 59,60. They serve as a stable support for the immobilization of nucleic acids and are efficient in removing non-specific interactions. One of the earliest studies utilizing the silica nanoparticles in ATP aptasensors was reported by Wang et al. 61. In their work, DNA probes are immobilized onto the silica nanoparticles and a Cy-5-tagged aptamer is hybridized to the immobilized DNA probes. The presence of ATP induces a conformational change of the aptamer-probe duplex when it binds to ATP, releasing the Cy-5-tagged aptamer to the solution and hence fluorescence is observed in the supernatant after separation. Upconversion nanoparticles (UCNPs) have also been used in the fluorescent ATP aptasensors. UCNPs have the advantage of low autofluorescence interference from biological samples as they are excited by low energy near-infrared photons, but emission happens in the visible region. In the work of He et al. 62, a split aptamer is used, whereby an ATP aptamer containing 27 nucleotides is split into two units. One unit modified with an amino group on one end, is immobilized onto the UCNPs. The other unit is tagged with a quencher BHQ1. In the presence of ATP, the quencher will be bought close to the UCNPs due to the formation of the aptamer-ATP complex. Hence, FRET occurs and the fluorescence of the UCNPs is quenched. In another report, quantum dots (QDs) were used as fluorescent reporters for ATP detection (Figure 4) 63. Streptavidin-conjugated 605QDs are first modified with 3'-biotin-tagged DNA strands. 3′-Cy5-labeled DNA strands are brought into close proximity to the QDs through capture DNA strands, in which the capture DNA strands have sequence complementary to the abovementioned two DNA strands. FRET takes place between the QDs and the 3′-Cy5-labeled DNA strands with fluorescence from Cy5 is observed at 670 nm. On the other hand, the presence of ATP releases the 3′-Cy5-labeled DNA strands because ATP preferentially binds to the aptamer in the capture DNA strands. As a result of ATP binding, FRET is disrupted and fluorescence from the QDs is observed at 605 nm. QDs have the advantages of high stability and long fluorescence lifetime compared to conventional fluorophores and easily modified to fit for purpose. However, QDs also suffer from the disadvantage of being toxic to human health.
Figure 4.
Schematic illustration of ATP detection by FRET principle. 3'-biotin-modified DNA is immobilized onto streptavidin-coated QDs. 3′-Cy5-labeled DNA is brought into proximity to the QDs through capture DNA. Fluorescence quenching occurs. The binding of ATP induces the release of 3′-Cy5-labeled DNA, causing a fluorescence wavelength change. (Reproduced with permission from 63. © 2008 Springer.)
Similar to nanoparticles being utilized in the labeled fluorescent ATP aptasensors, nanoparticles have also widely been studied for the construction of the non-labeled fluorescent ATP aptasensors. For instance, An ATP aptasensor was fabricated as follows: Probe DNA is immobilized onto silica nanoparticles and aptamer hybridizes to the probe DNA, forming a probe DNA-aptamer duplex. Hoechst33258 is used as a reporter, which intercalates to the probe DNA-aptamer duplex, giving fluorescence. Similar to many other fluorescent ATP aptasensors mentioned above, ATP detection is based on fluorescence quenching when ATP binds to the aptamer and releases the intercalated Hoechst33258 into solution 64. In addition, QDs in conjugation with fluorescence polarization immunoassay (FPIA) were explored for ATP detection 65. FPIA is based on the principle that the fluorescence polarization value will be higher when the fluorophore has a bigger volume (weight) and slower rotation. With this principle in mind, He et al. immobilized an aptamer onto CdS QDs. In the presence of ATP, the immobilized aptamer and another aptamer modified with an antigen (mouse anti-digoxin antigen) bind to ATP 65. The fluorescence signal is amplified when the antibody, anti-digoxin immobilized to gold nanoparticles binds to the entire complex, resulting in the formation of a very big conjugate and hence drastically lowers the rotation of the conjugate thus producing a much higher fluorescence polarization value. Detection limit was found to be 1000 times lower than that obtained without the gold nanoparticles.
Not limiting to nanoparticles, nanotubes such as single-walled carbon nanotubes (SWCNTs) have also received attention in ATP aptasensors. Covalent and non-covalent interactions of nucleic acids with SWCNTs were reported 66,67. Furthermore, SWCNTs are efficient fluorescence quencher, which makes them good candidates for uses in fluorescent biosensors. Several reports have studied on the non-covalent interactions of SWCNTs with biomolecules labeled with fluorophores or organic compounds 68-70. An example was reported by Zhang et al. on the detection of ATP based on SWCNTs (Figure 5) 71. An ATP aptamer is modified with a fluorophore - 6-carboxy-fluorescein (FAM). Without ATP, the FAM-labeled aptamer moves close to the SWCNTs because of π-stacking interaction, which quenches the fluorescence of FAM and also helps to disperses the SWCNTs in solution. Fluorescence is recovered when ATP competes against the SWCNTs for the labeled aptamer, disrupting the aptamer/SWCNT interaction. Although the non-covalent interaction approach eliminates additional steps to form covalent bonds between the aptamer and the SWCNTs, non-target molecules may easily interact with the aptamer and the SWCNTs. This is especially so when applied to a cellular environment containing numerous types of biomolecules. In addition, SWCNTs have the disadvantage of strong interaction with nucleic acids due to wrapping of nucleic acids around the nanotubes. Hence, the release of the labeled aptamer from the SWCNTs may be difficult, thereby causing less efficient sensing ability. Therefore, as very effective fluorescence quenchers, two-dimensional carbon nanomaterials such as graphene 72 and graphene oxide 73 have been applied in the construction of the fluorescent ATP aptasensors. Unlike SWCNTs, graphene oxide possesses intermediate strength of interaction with nucleic acids with well-balanced non-covalent interaction and desorption ability. This property makes graphene oxide attractive as a nanomaterial platform for ATP sensing. In the paper by Liu et al. 74, a comparison was made between covalent attachment and physisorption of an aptamer to graphene oxide. It was demonstrated that the fluorescence intensity is stronger for the covalent attached aptamer when applied to intracellular detection. The covalently attached aptamer to graphene oxide also has the potential to be regenerable for repeated use, making it attractive for ATP detection.
Figure 5.
Scheme for signaling ATP interaction by the assembly of SWCNTs and dye-labeled aptamer. (Reproduced with permission from 71. © 2010 Elsevier Ltd.)
Table 1 summarizes the analytical performance of fluorescent ATP aptasensors.
Table 1.
Analytical performance of fluorescent ATP aptasensors.
| Type of aptamer | Detection technique | Signal generation mechanism | Linear range | Detection limit | Ref |
|---|---|---|---|---|---|
| Molecular beacon-based aptamer | Fluorometry | Fluorescence recovery through disrupting Long range resonance energy transfer with background minimization by carbon nanotubes | 0.8-80 μM | 0.5 μM | 29 |
| Single-stranded aptamer | Fluorometry | Molecular beacon-assisted fluorescence recovery | 10-500 nM | 0.1 nM | 52 |
| Single-stranded aptamer | Fluorometry | Fluorescence sensing of a double-receptor sandwich supramolecule among the aptamer, ATO and a metal complex-based triphosphate receptor after separation | 0.2-5 nM | 37 pM | 53 |
| Hybridized aptamer | Fluorometry | Isothermal circular strand-displacement polymerization amplification | 1.0-10 nM | 0.26 nM | 54 |
| Single-stranded aptamer | Fluorometry | ATP-binding initiated ligation and RCA | 10-100 μM | 1.0 μM | 55 |
| Single-stranded aptamer | Fluorometry | ATP-binding triggered dislodge of fluorophore-labeled aptamer from graphene oxide | ---------- | 60 nM | 56 |
| DNAzyme containing aptamer | Fluorometry | DNAzyme- Modulated Growth of CdS QDs | 5-50 μM | 5.0 μM | 57 |
| Hybridized aptamer | Fluorometry | ATP-binding triggered liberation of DNAzyme | 10-80 nM 0.1 - 1.0 μM |
10 nM | 58 |
| Hybridized aptamer | Fluorometry | Dehybridization of fluorophore-tagged aptamer on silica nanoparticles through the formation of ATP-aptamer complex | 0.05-1.0 mM | 0.034 mM | 61 |
| Split aptamer | Fluorometry | FRET between silica upconversion nanoparticles and fluorophore-tagged split aptamer | 2-16 μM | 1.7 μM | 62 |
| Hybridized aptamer | Fluorometry | FRET between quantum dots and fluorophore-tagged split aptamer | 0.1-1 mM | 2.4 μM | 63 |
| Single-stranded aptamer | Fluorometry | Carbon nanotube-assisted recovery of fluorescence | 10-800 nM | 4.5 nM | 71 |
| Single-stranded aptamer | Fluorescence imaging | Fluorescence recovery of fluorophore-tagged aptamer immobilized on graphene oxide through the formation of ATP-aptamer complex | 0.125--2 mM | 0.019 mM | 74 |
| Split aptamer | Fluorometry | Silver nanocluster-assisted fluorescence turn on | 0.001--1 mM | 0.2 μM | 38 |
| Single-stranded aptamer | Fluorometry | Intercalation of light switching complex | 20-100 nM | 20 nM | 42 |
| Hybridized aptamer | Fluorometry | Metal-enhanced fluorescence of intercalator | 0.01-100 μM | 1.3 nM | 44 |
| Hybridized aptamer | Fluorometry | Intercalating dye with harvesting cationic tetrahedralfluorene | ---------- | 20 μM | 46 |
| Hybridized aptamer | Fluorometry | Fluorescence from intercalating dye after separation | 25-200 μM | 20 μM | 64 |
| Split aptamer | Fluorescence polarization | Gold nanoparticle-assisted fluorescent polarization | 8 pM-0.24 mM | 1.8 pM | 65 |
| Hybridized aptamer | Fluorescence imaging | Fluorescence turn on through conformational change after ATP binding on a sol-gel matrix | Low μM-mM | ---------- | 49,72 |
| Split aptamer | Fluorometry | Amplification by nuclease-assisted recycling of ATP | ---------- | 20 nM | 75 |
| Hybridized aptamer | Fluorometry | Fluorescence from Cu nanoclusters formed on ds-DNA | 0.05-500 μM | 28 nM | 76 |
| Hairpin aptamer | Fluorometry | Metal paired conformational switch and fluorescent gold nanoclusters | 100-2000 nM | 48 nM | 77 |
| Hairpin aptamer | Fluorometry | Structure switching of hairpin aptamers and exonuclease catalyzed target recycling | 10 nM-2 μM | 9.5 nM | 78 |
| Single-stranded aptamer | Fluorometry | Fluorescence of intercalating dye bound to ATP-aptamer complex coupled to exonuclease digestion | 0.5-50 μM | 140 nM | 79 |
| Hairpin aptamer | Fluorometry | Fluorescence turn on through conformational change after ATP binding | 50 nM-20 μM | 25 nM | 80 |
| Hairpin aptamer | Fluorometry | Fluorescence of intercalating dye bound to ATP-aptamer complex | --------- | 10 μM | 81 |
| Hybridized aptamer | Fluorescence anisotropy | Amplified detection through ATP-triggered hybridization chain reaction | 0.2-2 μM | 100 nM | 82 |
| Hairpin aptamer | Fluorometry | Fluorescence turn on through conformational change after ATP binding | up to 2.5 mM | 1.45 μM | 83 |
| Hybridized aptamer | Fluorometry | Fluorescence of metal complex bound to ATP-aptamer complex | 2.5-100 μM | 2.5 μM | 84 |
| Single-stranded aptamer | Fluorometry | Fluorescence recovery of fluorophore-tagged aptamer adsorbed on graphene upon ATP binding | 0.5-50 μM | 0.5 μM | 72 |
| Hybridized aptamer | Fluorometry | Fluorescence recovery of fluorophore-tagged aptamer adsorbed on Ag nanoparticles | 0-0.5 mM | 8 μM | 85 |
| Single-stranded aptamer | Fluorometry | Fluorescence recovery of fluorophore-tagged aptamer adsorbed on carbon nanotube upon ATP binding | 0.28-28 nM | 0.1 nM | 86 |
| Single-stranded aptamer | Fluorescence Anisotropy | Graphene oxide-assisted fluorescence anisotropy | 0.5-250 μM | 0.12 μM | 73 |
| Hybridized aptamer | Fluorometry | ATP binding-triggered strand displacement amplification | ---------- | 20 nM | 87 |
| Hybridized aptamer | Fluorometry | Aptamer/quantum dot based resonance energy transfer | 0.1 nM-1 μM | 0.01 nM | 88 |
| Hybridized aptamer | Fluorometry | fluorescence quenching by graphene oxide upon ATP binding | 3-320 μM | 0.45 μM | 89 |
| Split aptamer | Fluorometry | Formation of excimer in cyclodextrin upon ATP binding | 0.5-50 μM | 80 nM | 90 |
| Split aptamer | Fluorometry | Formation of fluorescent molecules through click chemistry upon ATP binding | 0.03-100 nM | 0.02 nM | 91 |
| Quencher-free aptamer | Fluorometry | Fluorescence turn-on upon ATP binding | 10 -50 mM | 2.0 mM | 92 |
| Hybridized aptamer | Fluorometry | ATP binding-triggered exonuclease-assisted target recycling | 0.1 nM-50 μM | 43 pM | 93 |
| Hairpin aptamer | Fluorescence anisotropy | ATP binding-induced Fluorescence anisotropic change | 1.0-200μΜ | 1.0 μM | 94 |
| Hybridized aptamer | Fluorometry | Fluorescence quenching of intercalating dye upon ATP binding | 100 nM-10 mM | 14 nM | 45 |
| Single-stranded aptamer | Fluorometry | ATP-binding-triggered fluorescence turn-on of silver nanoclusters | 0.50-8.0 μM | 92 nM | 38 |
| Hybridized aptamer | Fluorometry | ATP-binding-initiated aptamer molecular machine of target recycling | 50-500 nM | 25 nM | 95 |
2.4. ATP aptamer-based fluorescence imaging
In addition to the fluorescent ATP aptasensors mentioned above, ATP aptamers have also found applications in in vivo imaging of ATP 96-104. The first attempt of imaging ATP in living cells using ATP aptamers was described by Mirkin's group in which ATP aptamer/fluorescent reporter duplex-coated gold nanoparticles were utilized to visualize ATP in living cells 96. The fluorescence of the reporters is effectively quenched by the gold nanoparticles due to the close proximity between the fluorescent reporters and the gold nanoparticles. Upon binding to ATP the fluorescent reporters are dislodged from the gold nanoparticles due to conformational changes of the ATP aptamers and the fluorescence of the reporters is restored. The fluorescence intensity of the dislodged reporters can be used to quantify ATP in cells. Later, it was shown that the gold nanoparticles loaded with ATP aptamer can be configured to display both ATP and pH responsive probes to image ATP in lysosomes at the subcellular level 97. More recently, instead of gold nanoparticles, gold nanocages carrying positive charges on their surface were utilized to load anionic ATP aptamers via electrostatic interaction 98. Upon binding to ATP inside living cells, the aptamers are desorbed from the gold nanocages and entrapped fluorophores are released, thus generating a fluorescence image.
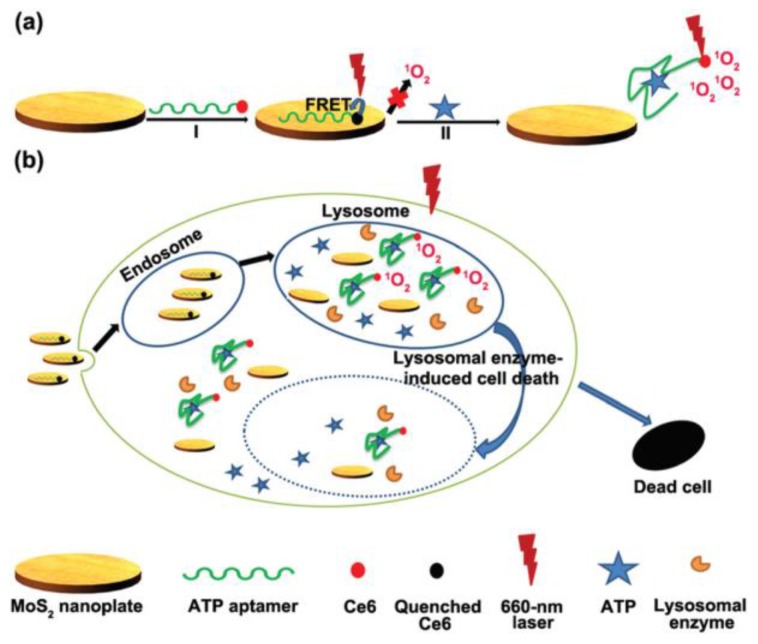
In another attempts, graphene oxide nanosheets were involved in the construction of ATP imaging probes instead of gold nanoparticles 100-103. Strong interaction between ATP aptamer-containing fluorescent probes and the graphene oxide nanosheets effectively quenched the fluorescence of the probes. Again, the fluorescence of the probes is restored upon binding to ATP which results a conformational change. It was shown that multiple aptamer sensing platforms can be constructed by a judicious design of multiple aptamer-tagged probes. Other two-dimensional nanomaterials like MoS2 nanoplates were also tested as carriers for the construction of a photodynamic agent chlorine e6 (Ce6)-labeled ATP aptamer for cell imaging and photodynamic therapy (Figure 6) 104. It was shown that in addition to visualizing intracellular ATP, the labeled aptamer released from the MoS2 nanoplates initiates the production of reactive oxygen species upon laser irradiation which consequently induces cell death by a lysosomal pathway.
Figure 6.
(a) Schematic representation of the Ce6-tagged ATP aptamer loaded MoS2 nanoprobes for reactive oxygen species upon binding to ATP, (b) endocytosis of the nanoprobes for ATP imaging and ATP-mediated production of reactive oxygen species upon laser irradiation. (Reproduced with permission from 104. © 2015 Royal Society of Chemistry)
Although encouraging results have been obtained in imaging ATP in living cells, there are some critical questions that we need to answer before we can take full advantage of ATP aptamers. Unfortunately, we do not have highly specific aptamers for ATP. The coexistence of ADP and AMP with ATP inside cells poses a great threat to ATP imaging. There is an urgent need to thoroughly investigate the specificity of ATP aptamers and to search for truly ATP-specific aptamers since all ATP aptamers developed so far show a considerable degree of affinity to ADP and AMP. The results remain disputable until ATP-specific aptamers are developed. We also need to fully understand the effect of complex biological matrices on the performance of ATP aptamers in cells. In addition, it will be more interesting if both ATP and ADP can be imaged simultaneously so as to provide dynamic pictures of the transformations between ATP and ADP in living organisms.
3. Colorimetric ATP aptasensors
Colorimetric detection is the visual observation or spectrometric measurement of a color change. It has the advantages of low cost, simplicity, and portability. Gold nanoparticles, hemin-G-quadruplex aptamers, and supramolecule-based aptamers are mainly used in the colorimetric ATP aptasensors. In addition to visual inspection, UV-vis spectrometry is often employed for quantitative determination of ATP.
3.1. Nanomaterial-assisted colorimetric ATP aptasensors
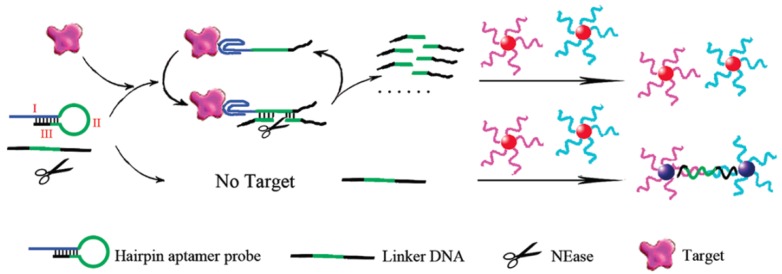
Contrary to fluorescent biosensors, gold nanoparticles have attracted much more attention in the development of colorimetric biosensors because their ultrahigh extinction coefficient (108-109) of the surface plasmon resonance absorption offers an excellent opportunity for the development of highly sensitive biosensors 105,106. Therefore, gold nanoparticles have been of great interest for uses in colorimetric detections of biomolecules including ATP due to their simplicity and ultrahigh extinction coefficient with a strong dependence on inter-particle distance 107. Distance dependent optical property results in a significant shift in UV-vis absorption or/and a distinct color change. Hence, approaches that can alter the distance among the gold nanoparticles upon target binding have been investigated in the colorimetric ATP aptasensors. A simple signal generation strategy relies on the alteration of the protective effect of nucleic acids on gold nanoparticles [108.109]. To significantly enhance the sensitivity of colorimetric ATP aptasensors, various amplification strategies have been proposed. One of the approaches is to modify the gold nanoparticles with DNA and the use of nuclease 110. As reported in the work by Li et al., a cyclic enzymatic signal amplification strategy was used to improve the detectability of a colorimetric aptasensor (Figure 7) 110. Gold nanoparticles are first modified with two types of DNA strands, each type complementary to one end of a linker DNA. A hairpin aptamer is also prepared. In the absence of ATP, the linker DNA induces the aggregation of the gold nanoparticles by hybridization with the two types of DNA strands immobilized on the gold nanoparticles. However, with ATP added, the hairpin aptamer opens up and forms the aptamer-ATP complex, which could subsequently attach to the linker DNA. This binding then induces nicking endonuclease to break the linker DNA into two fragments. As a result, the cleaved linker DNA fragments are released and the freed-up aptamer-ATP complex repeats the cycle of binding to another linker DNA. The cleaved linker DNA is unable to induce the aggregation of the gold nanoparticles through hybridization. Hence, a red color is observed instead of the blue color when ATP is absent. This aptasensor produced a low detection limit of 100 nM by the naked eye as the regeneration process allows a very large amount of the linker DNA to be cleaved in presence of a small amount of ATP. Another example of utilizing gold nanoparticles in the construction of the colorimetric ATP aptasensors was described by Zhao and co-workers 111.They proposed a simple and rapid colorimetric ATP aptasensor that exploits structure-switching ATP aptamers and salt-induced aggregation of gold nanoparticles.
Figure 7.
Schematic illustration of the colorimetric detection of ATP based on cyclic enzymatic signal amplification. (Reproduced with permission from 110. ©2102 American Chemical Society)
Since the preparation of DNA-modified gold nanoparticles is rather time-consuming, usually requiring a multistep procedure, alternative approaches using unmodified gold nanoparticles were explored. The concept of using unmodified gold nanoparticles relies on the difference in electrostatic interaction between single-stranded (ss-DNA) and double-stranded (ds-DNA) DNA with gold nanoparticles. Gold nanoparticles are stabilized by negative ions, whereby the repulsion between the adsorbed negatively charged ions prevent the gold nanoparticles from aggregation. The addition of salt minimizes the electrostatic repulsion and induces aggregation. The relatively rigid double helix structure of ds-DNA largely conceals its nucleobases and exposes its negatively charged phosphate backbone; hence prevents the adsorption of the ds-DNA on the gold nanoparticles. On the other hand, being uncoiled, ss-DNA has its nucleobases exposed. Van der Waals interaction becomes dominant and the ss-DNA could get close and coil around the gold nanoparticles. The adsorption of ss-DNA strands effectively stabilizes the gold nanoparticles and prevents aggregation in the presence of salt 112. Based on this principle, Wang et al. proposed an aptamer-target binding readout technique for the detection of ATP 113. First, salt is added to induce the aggregation of the gold nanoparticles and an aptamer bound to its complementary sequence is added too, thus producing a blue colored solution of aggregated gold nanoparticles. The presence of ATP triggers the formation of the aptamer-ATP complex and releases the complementary sequence to coil around the gold nanoparticles to block the salt-induced aggregation. Consequently, instead of a blue colored solution a red colored solution is observed due to the non-aggregated gold nanoparticles in the presence of ATP.
3.2. Amplified colorimetric ATP aptasensors
Nucleic acid sequences which have many guanine bases at specific locations, can interact through strong hydrogen bonds to form a four-stranded helical structure - G-quadruplex. Also, the G-quadruplex can selectively interact with hemin, forming a catalytic G-quadruplex-hemin complex - DNAzyme. This DNAzyme acts like peroxidase to catalyze the oxidation of hydrogen peroxide-mediated reactions 114. The oxidation of peroxidase substrates such as 3,3',5,5'-tetramethylbenzidine (TMB) or 2,2'-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid (ABTS2-) catalyzed by the DNAzyme produces a color change. This principle was applied to ATP detection by incorporating the G-quadruplex sequence in an aptamer 115. For example, gold nanoparticles are immobilized onto peptide nanospheres to form a nanocomposite. The G-quadruplex containing aptamer modified with thiol then binds to the nanocomposite. Another aptamer is immobilized onto a 96-well plate. The presence of ATP will bind to both aptamers on the nanocomposite and inside the well. This brings the nanocomposite containing the G-quadruplex into the well. After complexing with hemin, catalytic oxidation of TMB occurs, giving a yellow color to the solution. UV-vis spectrometry was further used to evaluate the performance of this aptasensor. A detection limit of 1.35 pM was obtained.
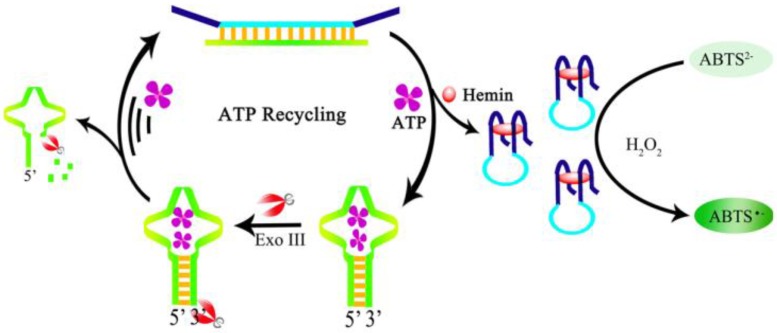
Moreover, to increase the sensitivity, a color enhancement strategy by an amplification process through ATP recycling was developed by Gong et al. 116. A ds-DNA sequence is prepared containing the G-quadruplex sequence and the ATP aptamer sequence. The ds-DNA has four single-stranded termini hence resists the digestion by exonuclease Exo-III. Exo-III can only cleave sequences with blunt ends or recessed 3' termini. In the presence of ATP, the aptamer is dehybridized and forms a hairpin structure with a blunt end upon binding to ATP, which in turn induces cleavage by Exo-III to release the bound ATP and triggers a target recycling process. The dehybridization and Exo-III cleavage also release the G-quadruplex sequence. After the formation of the DNAzyme and in the presence of H2O2, colorless ABTS2- is oxidized to a green colored product ABTS- (Figure 8). A very large amount of hemin-G-quadruplex complex could be formed through the ATP regeneration process by the Exo-III cleavage. Consequently, the sensitivity of the aptasensor is significantly improved with negligible interference from other nucleotide triphosphates. Likewise, amplified colorimetric detection of ATP can be realized by self-assembled aptamer constructs that activate a DNAzyme cascade 117.
Figure 8.
Schematic illustration of ATP recycling through Exo-III cleavage and DNAzyme formation. (Reproduced with permission from 116. ©2104 Elsevier Ltd)
4. Chemiluminescent ATP aptasensors
Chemiluminescence involves a chemical reaction producing one or more reaction products existing in an electronically excited state, which eventually returns to the ground state through the emission of light 118. The intensity of the light emitted is directly associated with the reactants and the catalyst of the reaction. Unlike the fluorescent or colorimetric technique where a light source and complicated optics are required, the main advantage of using chemiluminescence as the detection technique in ATP aptasensors is its simplicity of instrumentation and high sensitivity due to a practically zero background 118. There are, however, some limitations which should be noted when considering its use. For example, there are not many chemiluminescent compounds available, hence the technique is only limited to a relatively small number of analytes. Fortunately, for the detection of ATP, a well-known chemiluminescent platform-luminol-H2O2 system can be conveniently adopted for the construction of the chemiluminescent ATP aptasensors.
4.1. Chemiluminescent ATP aptasensors involving chemiluminescence resonance energy transfer (CRET)
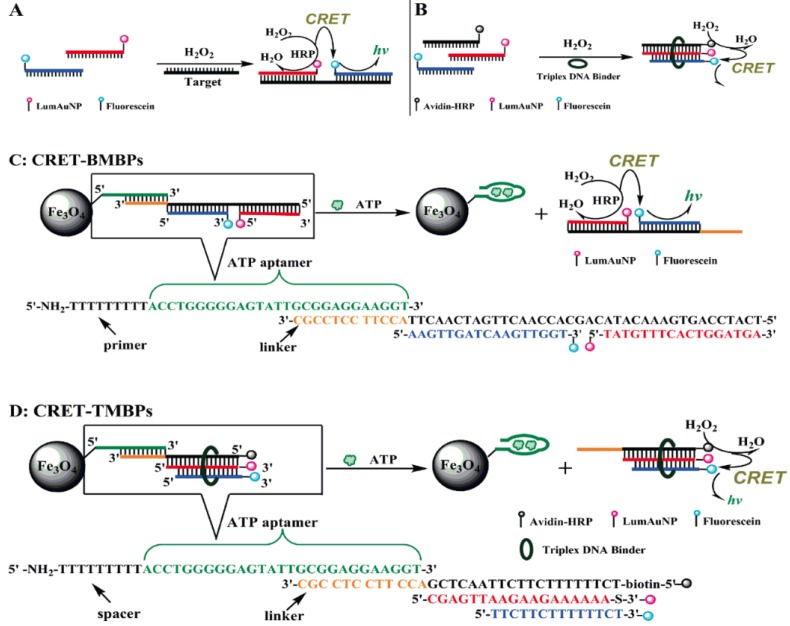
Huang et al. suggested the exploitation of CRET technique for the construction of chemiluminescent biosensors 119. CRET involves the non-radiative energy transfer from a chemiluminescent donor to a suitable acceptor. Soon after, Zhang et al. reported a chemiluminescent ATP aptasensor inspired by the concept of CRET using a luminol-H2O2-horseradish peroxidase-fluorescein system (Figure 9) 120. In this aptasensor, fluorescein is able to absorb part of the energy (~425 nm) of the excited luminol and subsequently re-emit it at a longer wavelength (~510 nm). Based on this CRET process, sensing probes termed as CRET-BMBP and CRET-TMBP are designed. First, an ATP-specific aptamer is attached to the surface of magnetic nanoparticles and the linking region in this aptamer has an extension that can hybridize with CRET-BMBP or CRET-TMBP probes, respectively. The reason of utilizing the magnetic nanoparticles is that the separation of reacted probes from unreacted ones can be conveniently achieved by simply using a permanent magnet. When ATP is present, the aptamer switches to its preferred aptamer-ATP structure from the aptamer-linker conformation. This results in the dissociation of CRET-BMBP or CRET-TMBP probes from the aptamer. Since the chemiluminescence intensity detected is due to the CRET-BMBP or CRET-TMBP probes. The CRET signal is therefore proportional to the amount of ATP. The proposed CRET-based aptasensor offered detection limits of 0.11 µM and 0.32 µM for ATP by BMBP and TMBP, respectively. This aptasensor has been successfully applied to the detection of ATP in leukemia and breast cancer cells. Similarly, leveraging on CRET, ATP detection can be realized by measuring the fluorescent intensity of QDs excited by CRET between luminol and the QDs 121. It was shown that the fluorescence intensity increases linearly with the concentration of ATP from 50 to 230 µM with a detection limit of 0.185 µM and a selectivity factor of ~20 against ATP analogues such as thymidine triphosphate (TTP), CTP, and GTP.
Figure 9.
Scheme of the CRET-signal conjugates and the application of CRET in the determination of ATP. (Reproduced with permission from 120. ©2009 American Chemical Society)
The concept of CRET was also applied to the detection of ATP involving a DNAzyme-aptamer conjugate 122. Due to its peroxidase-like activity, the DNAzyme effectively catalyzes the oxidation of luminol in the presence of H2O2 to generate chemiluminescence. A conformational change brought about by the binding of ATP to the aptamer facilitates CRET to occur by bring the quencher close the DNAzyme. In another report, Strands of ss-DNA consist of both DNAzyme and ATP aptamer segments were utilized for the detection of ATP 123. In the presence of ATP and hemin, The DNA strands selectively bind to ATP and self-assemble into the peroxidase-like DNAzyme structure (hemin-G-quadruplex complex). Chemiluminescent ATP detection is realized by tapping in the peroxidase property of the DNAzyme. Again, CRET was incorporated to further develop the sensing platform 123.
4.2. Nanomaterial-assisted chemiluminescent ATP aptasensors
Leveraged on the easy use of magnetic beads in the separation of reactants and products, Li et al. proposed a chemiluminescent ATP aptasensor based on magnetic nanoparticle-assisted strand displacement amplification and target recycling 124. It was reported that the light intensity emitted from the luminol-H2O2-horseradish peroxidase system is linearly correlated to ATP concentration from 1.0 to 10 nM with a detection limit of 0.61 nM. Furthermore, as with the fluorescent ATP aptasensors, nanomaterials have also found applications in the chemiluminescent ATP aptasensors. For example, a chemiluminescent aptasensor was developed for the detection of ATP using graphene oxide, relying on the preferential binding of graphene oxide to ss-DNA over ATP-aptamer complex and the instantaneous derivative reaction between phenylglyoxal and guanosine in the aptamer strand adsorbed on the graphene oxide. The addition of ATP weakens the interaction between the aptamer and graphene oxide and results in a significant reduction of the chemiluminescence signal. A detection limit of 1.4 nmol with good selectivity (≥5 folds) against CTP, GTP, and UTP was obtained. Nonetheless, the rather limited options in the configuration of chemiluminescence platforms severely obstruct the progress of chemiluminescent ATP aptasensors.
5. Other optical ATP aptasensors
Other than the discussed techniques of fluorometry, colorimetry, and chemiluminescence, there are also other optical techniques used in ATP aptasensors like localized surface plasmon resonance (LSPR), surface-enhanced Raman spectroscopy (SERS), and resonance light scattering (RLS). Nonetheless, these optical techniques are not as popular as fluorometry and colorimetry. In the following section, the current status of applying the above-mentioned optical techniques will be reviewed.
5.1. LSPR ATP aptasensors
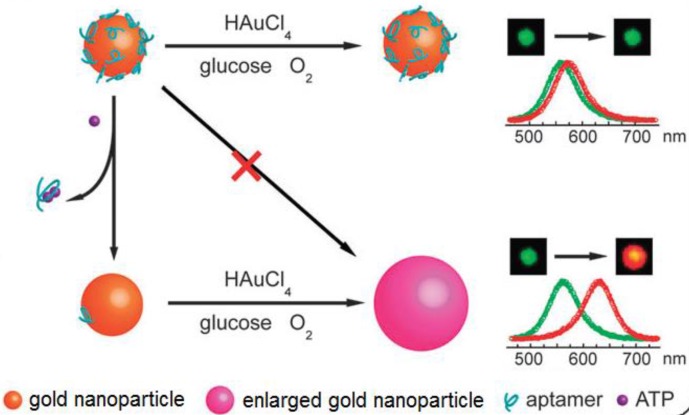
LSPR is a result of a coherent oscillation of electrons taking place on the surface of a nanostructured material (mainly gold and silver) in response to an alternating electromagnetic field of incident light 125. LSPR, extremely sensitive to the geometry (size, shape, and pattern) and local dielectric environment of the nanostructure, has recently been applied to ATP aptasensors 126,127. For example, Liu et al. proposed an ATP aptasensor based on aptamer-regulated self-catalytic growth of single gold nanoparticles (Figure 10) 126. It was observed that the presence of ATP aptamer on the surface of the gold nanoparticles effectively suppresses the glucose oxidase-like catalytic activity of the gold nanoparticles, which in turn, inhibits the growth of the gold nanoparticles. On the other hand, the formation of the aptamer-ATP complex detaches the aptamer from the gold nanoparticles and recovers their catalytic activity. Consequently, the gold nanoparticles are enlarged in a growth medium containing HAuCl4 and glucose, thus producing an observable shift in LSPR.
Figure 10.
Schematic illustration of the LSPR aptasensor. The plasmonic signal is generated by the ATP-induced conformational change of surface adsorbed ATP aptamer that recovers the self-catalytic activity of the gold nanoparticles. (Reproduced with permission from 126. © 2012 Royal Society of Chemistry)
Cheng et al. applied a split aptamer and gold nanoparticles for the detection of ATP using LSPR 127. The two parts of a split aptamer are co-immobilized on the same gold nanoparticles. The split aptamer on the gold nanoparticles can be reassembled into intact folded structure in the presence of ATP with two potential assembly types and both of them induce the assembly of gold nanoparticle-DNA conjugates, thus giving rise to a measurable change in LSPR. The detection limit of this LSPR aptasensor was estimated to be 24 μM. The main limitation of the LSPR ATP aptasensors is that many times the refractive index change may not be significant enough for a measurable signal to be obtained at low concentrations 128.
5.2. SERS ATP aptasensors
Raman spectroscopy has been commonly used to characterize molecules based on their unique capability of detecting the interaction of light with molecules 129. However, the most common problem encountered in the application of Raman spectroscopy for small biomolecules is their low scattering cross-sections and weak signals. Fortunately, the development of SERS opened up new prospects for Raman spectroscopy. The generally accepted mechanisms of SERS are the electromagnetic mechanism and chemical mechanism 129,130. SERS is a surface-sensitive technique that enhances Raman scattering of molecules adsorbed on nanostructured metal surfaces 131. SERS has since evolved into a very powerful technique in the field of analytical chemistry especially at single molecule levels 132,133. In addition, SERS is a nondestructive and highly sensitive technique which requires minimal sample preparation.
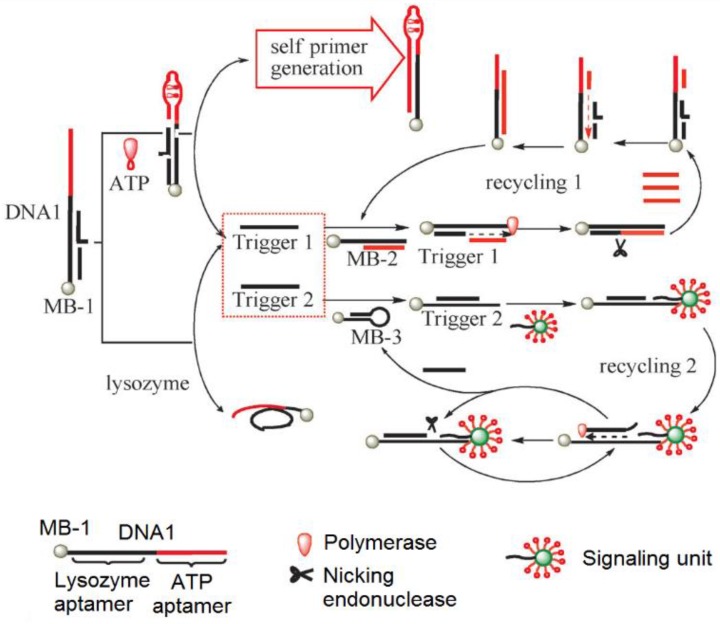
To develop SERS-based ATP aptasensors, a primer self-generation strand-displacement polymerization (PS-SDP) was used to construct a bifunctional aptasensor for the detections of ATP and lysozyme (Figure 11) 134. With the cooperation of a nicking endonuclease and a polymerase, amplified detections of ATP and lysozyme were achieved through a two-cycle replication-scission displacement mode. As shown in Figure 11, based on a structure-switching aptamer, three types of functional components, namely a target recognition element DNA1, a transduction component DNA2, and a signaling unit (gold nanoparticle-based SERS bio-barcodes probe), are involved in the amplification process. DNA1 immobilized on magnetic bead 1 (MB-1) binds trigger 1 and trigger 2. DNA2 binds to a hairpin DNA immobilized on MB-2 and DNA3 immobilized on MB-3. The signaling unit acts both as the primer for the polymerization and as the reporter for Raman signal generation. In the presence of ATP, the ATP aptamer is triggered to undergo structure switching to form a stem-loop structure, initiating the PS-SDA process. Consequently, amplified SERS detection of ATP in the range of 0.05 to 10 nM was realized by measuring the Raman signal of SERS bio-barcode probes attached to hairpin DNA on MB-3. In addition, the Raman enhancement effect of a gold nanotip array and the feasibility of constructing a gold nanotip-based ATP aptasensor were thoroughly studied by Jiang et al. 135. It was observed that picomolar sensitivity for ATP could be realized owing to the high local electromagnetic field around each of the individually addressable nanotips of the array.
Figure 11.
Schematic representation of the PS-SDP-based bifunctional SERS ATP aptasensor. (Reproduced with permission from 134. © 2013 Willey-VCH)
5.3. RLS ATP aptasensors
RLS is based on the self-assembly or aggregation of nanomaterials such as gold nanoparticles 136,137. It displays the advantages of simplicity, speed, and high sensitivity. This technique has been widely used in the determination of analytes, such as inorganic ions 138, drugs 139, and biological macromolecules 140.
As mentioned earlier, unlike ss-DNA, ds-DNA is unable to protect gold nanoparticles against salt-induced aggregation due to it high rigidity, concealed nucleobases, and largely exposed negative charges. On the other hand, there is a distinct difference in RLS between homogeneously dispersed and aggregated gold nanoparticles. Based on these two properties, a simple RLS ATP aptasensor was fabricated by building an ATP aptamer sequence into one of the two strands of a duplex DNA 141. Upon binding to ATP, a stable aptamer-ATP complex is formed and consequently a ss-DNA strand is released. As a result, the gold nanoparticles are protected from aggregation when NaCl is added to the solution, whereas unprotected gold nanoparticles are aggregated, thus displaying a resonance scattering peak at 590 nm with a linear calibration curve between 6.6 and 110 nM. To further increase the sensitivity of the RLS aptasensors, a cuprous oxide nanoparticle formation process is incorporated in the ATP sensing procedure, taking advantage of the catalytic effect of the DNA-protected gold nanoparticles on the formation of the cuprous oxide nanoparticles in a mixture of glucose and cupric ions. This additional amplification step successfully lowered the calibration curve to 2.2-220 nM with a detection limit of 0.5 nM.
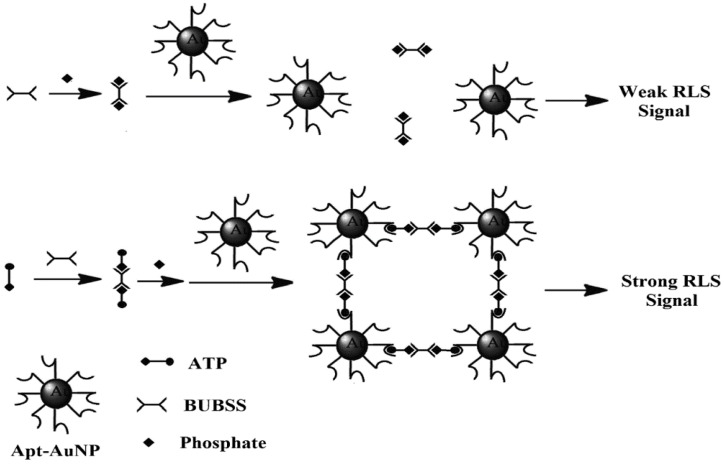
In a more recent paper, RLS was exploited for direct detection of ATP in aqueous solutions in the absence of any prior separation procedure 142. The materials involved were a binuclear uranyl complex - bis-uranyl-bis-sulfosalophen (BUBSS) and ATP aptamer-coated gold nanoparticle (Apt-AuNP) probes. Previous studies have shown that uranyl-salophen complexes could bind strongly to phosphate groups with high selectivity 143,144. ATP, the target of interest containing three phosphate groups, is an excellent analyte for RLS. On the other hand, the gold nanoparticles modified with the ATP aptamer function as a capturing agent for RLS. The sensing principle is as follows: In the presence of ATP, a specific reaction occurs between BUBSS and two ATP molecules to form a linear supramolecule. Consequently, one supramolecule triggers the binding of two gold nanoparticles, thus leading to the aggregation of the gold nanoparticles and producing a strong RLS signal (Figure 12). Under optimal conditions, a linear correlation between the RLS signal and the concentration of ATP was obtained from 5 to 500 nM with a detection limit of 4.5 nM. Selectivity tests showed that ATP gives a RLS signal which is at least 10 times higher than those obtained with its analogues such as GTP, CTP, UTP, adenosine, as well as AMP and ADP. Satisfactory results were also obtained when the proposed aptasensor was applied to the analysis of ATP containing drugs and urine samples.
Figure 12.
Diagram showing the reaction of ATP with BUBSS and Apt-AuNPs. (Reproduced with permission from 142. ©2015 Springer.)
6. Challenges and opportunities
Despite impressive progress in the research and development of the optical ATP aptasensors, none of them has clearly represented a fundamental breakthrough, becoming a viable alternative to the conventional bioluminescence method. Future research efforts should, therefore, be dedicated to the development of the optical ATP aptasensors with much improved sensitivity, selectivity, and robustness without engaging complicated fabrication and detection protocols. One possible approach is to couple nanomaterials to single-step amplification schemes and highly affinitive and selective aptamers to a simple signal readout mechanism such as litmus test or colorimetry. The excellent optical properties of nanomaterials may offer a convenient means for sensitivity improvement 145.
On the other hand, although displaying several advantages like the simplicity of synthesis, versatility in labeling and derivatization, and excellent stability, ATP aptamers with high affinity and specificity still remain rare. Increasing evidence has indicated that most of the aptamers are still not legitimate competitors to their antibody counterparts in term of affinity and specificity. As for ATP aptamers, a number of ATP aptamers with varied sequences and reasonable affinity towards ATP have been discovered since the invention of SELEX and in vitro selection for aptamer screening. Unfortunately, their poor selectivity towards other adenosine phosphate like ADP and AMP largely limits their applications in the analysis of real-world biological samples since ADP coexists with ATP in most biological samples. Because most of the known ATP aptamers, if not all, show relatively high affinity towards adenine, but not to the sugar and the triphosphate moieties, and especially the triphosphate moiety, they also show similar affinities towards ATP, ADP, and AMP 146. This may also explain why TTP, CTP, GTP, and UTP instead of ADP and AMP were often used as potential interferents in selectivity studies of many of the optical ATP aptasensors mentioned above.
Although some encouraging results were reported in imaging ATP in cultured cells and in detecting ATP in samples with complicated biological matrices like cell lysate, progress along this direction has been slow. Evidently, the high complexity of biological samples together with the coexistence of considerably high levels of other adenosine phosphates, namely ADP and AMP, ferociously hinders any meaningful applications of the optical ATP aptasensors due chiefly to their low selectivity. Consequently, a grand challenge in ATP aptasensors is to significantly enhance their selectivity for ATP, approaches and techniques that are able to significantly enhance the specificity of the ATP aptamers or the ATP aptasensors are both fundamentally and technically critical. Future research efforts should be directed to significantly enhancing the affinity and specificity of aptamers and the ATP aptasensors. One plausible approach is to optimize the structure of the aptamer to enable its interaction with both the nucleobase and triphosphate moieties. Encouraging results have been obtained but with an unfortunate dilemma - the aptamer does show some degree of interaction with the triphosphate but with reduced affinity towards the nucleobase 147. Strong repulsive interaction between the triphosphate moiety of ATP and the anionic backbone of the aptamer may be formidable to overcome in the design of highly affinitive ATP aptamers. To circumvent this problem, one possible solution could be the use neutral nucleic acid analogues such as peptide nucleic acids or morpholinos. Another possible solution could be the use of modified nucleic acids instead of natural nucleic acids.
In addition, leveraging on the charge density difference between ATP and other adenosine triphosphates, improved selectivity towards ATP could be realized by introducing a secondary recognition mechanism that is capable of strongly interacting with the triphosphate moiety of ATP, for example, uranyl-salophen complex 53,143,144. Indeed, promising results have been observed in a more recent report by employing a cationic polymer to discriminate ATP from ADP and AMP 148. It was observed that the strong interaction between the triphosphate moiety of ATP and surface-confined cationic polymer effectively enriches ATP molecules, thus leading to improved selectivity. Meanwhile, research in the applications of aptamers in the optical ATP aptasensors should focus on enhancing the robustness of the aptasensors and on the design of secondary recognition mechanism and powerful amplification schemes to significantly enhance the sensitivity to offload interference from co-existing species, particularly AMP and ADP in biological samples.
7. Conclusions and prospects
Since the discovery of aptamers in the 1990s, a number of aptamers with reasonably high affinity have been discovered via highly efficient and fully automated SELEX processes. This is in drastic contrast to the much more laborious, time-consuming, and expensive procedures required to produce antibodies. The excellent chemical and biological stability of aptamers and high manipulability together with their biologically non-toxic nature give a clear advantage in the context of biomedical applications over their antibody counterparts.
The fast advent of aptamer research has heralded a new chapter in biosensors. The proof-of-concept research so far has already confirmed that ATP aptasensors are able to contribute to the advances of biosensors and clinical science. It is believed that aptamers will play an increasing role in analytical and clinical science in the near future. As compared to other types of popular aptasensors, and electrochemical ATP aptasensors in particular, each of these two kinds of ATP aptasensors has its own strengths and weaknesses. For instance, in addition to delivering comparable or better sensitivity, being heterogeneous in nature, electrochemical ATP aptasensors may offer a convenient means to minimize matrix effect by simply conducting ATP detection in a different medium after ATP has been captured by its aptamer. On the other hand, aptamer-based optical ATP sensing is mostly conducted in a homogeneous or near homogeneous manner, better reproducibility is often observed. More advantageously, optical ATP sensing can in principle be performed in-situ, in vivo, and sometimes in real-time such as ATP fluorescence imaging. In view of the rapid progress made in a relatively short period of time, it seems clear that the future of the optical ATP aptasensors remains promising.
Acknowledgments
This work is supported by Ministry of Education (Grant No. MOE2014-T2-081).
References
- 1.Cook SP, Vulchanova L, Hargreaves KM. et al. Distinct ATP receptors on pain-sensing and stretch-sensing neurons. Nature. 1997;387(6632):505–508. doi: 10.1038/387505a0. [DOI] [PubMed] [Google Scholar]
- 2.Cockayne DA, Hamilton SG, Zhu QM. et al. Urinary bladder hyporeflexia and reduced pain-related behaviour in P2X 3-deficient mice. Nature. 2000;407(6807):1011–1015. doi: 10.1038/35039519. [DOI] [PubMed] [Google Scholar]
- 3.Lowe JE, Cummings RG, Adams DH. et al. Evidence that ischemic cell death begins in the subendocardium independent of variations in collateral flow or wall tension. Circulation. 1983;68(1):190–202. doi: 10.1161/01.cir.68.1.190. [DOI] [PubMed] [Google Scholar]
- 4.Souslova V, Cesare P, Ding Y. et al. Warm-coding deficits and aberrant inflammatory pain in mice lacking P2X 3 receptors. Nature. 2000;407(6807):1015–1017. doi: 10.1038/35039526. [DOI] [PubMed] [Google Scholar]
- 5.Gourine AV, Llaudet E, Dale N. et al. ATP is a mediator of chemosensory transduction in the central nervous system. Nature. 2005;436(7047):108–111. doi: 10.1038/nature03690. [DOI] [PubMed] [Google Scholar]
- 6.Vlaskovska M, Kasakov L, Rong W. et al. P2X3 knock-out mice reveal a major sensory role for urothelially released ATP. J Neurosci. 2001;21(15):5670–5677. doi: 10.1523/JNEUROSCI.21-15-05670.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kinnamon SC, Finger TE. A taste for ATP: neurotransmission in taste buds. ATP-gated P2X receptors in health and disease. Front Cell Neurosci. 2013;7:264. doi: 10.3389/fncel.2013.00264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Finger TE, Danilova V, Barrows J. et al. ATP signaling is crucial for communication from taste buds to gustatory nerves. Science. 2005;310(5753):1495–1499. doi: 10.1126/science.1118435. [DOI] [PubMed] [Google Scholar]
- 9.Butterfield SM, Waters ML. A designed β-hairpin peptide for molecular recognition of ATP in water. J Am Chem Soc. 2003;125(32):9580–9581. [Google Scholar]
- 10.Zhao M, Wang M, Liu H. et al. Continuous on-site label-free ATP fluorometric assay based on aggregation-induced emission of silole. Langmuir. 2008;25(2):676–678. doi: 10.1021/la803762b. [DOI] [PubMed] [Google Scholar]
- 11.Mizukami S, Nagano T, Urano Y. et al. A fluorescent anion sensor that works in neutral aqueous solution for bioanalytical application. J Am Chem Soc. 2002;124(15):3920–3925. doi: 10.1021/ja0175643. [DOI] [PubMed] [Google Scholar]
- 12.Leach FR. ATP determination with firefly luciferase. J Appl Biochem. 1981;3:473–517. [Google Scholar]
- 13.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249(4968):505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 14.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346(6287):818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 15.Sassanfar M, Szostak JW. An RNA motif that binds ATP. Nature. 1993;364(6437):550–553. doi: 10.1038/364550a0. [DOI] [PubMed] [Google Scholar]
- 16.Huizenga DE, Szostak JW. A DNA aptamer that binds adenosine and ATP. Biochemistry. 1995;34(2):656–665. doi: 10.1021/bi00002a033. [DOI] [PubMed] [Google Scholar]
- 17.Lin CH, Patei DJ. Structural basis of DNA folding and recognition in an AMP-DNA aptamer complex: distinct architectures but common recognition motifs for DNA and RNA aptamers complexed to AMP. Chem Biol. 1997;4(11):817–832. doi: 10.1016/s1074-5521(97)90115-0. [DOI] [PubMed] [Google Scholar]
- 18.Penchovsky R, Breaker RR. Computational design and experimental validation of oligonucleotide-sensing allosteric ribozymes. Nat Biotechnol. 2005;23(11):1424–1433. doi: 10.1038/nbt1155. [DOI] [PubMed] [Google Scholar]
- 19.Sreedhara A, Li Y, Breaker RR. Ligating DNA with DNA. J Am Chem Soc. 2004;126(11):3454–3460. doi: 10.1021/ja039713i. [DOI] [PubMed] [Google Scholar]
- 20.Seetharaman S, Zivarts M, Sudarsan N. et al. Immobilized RNA switches for the analysis of complex chemical and biological mixtures. Nat Biotechnol. 2001;19(4):336–341. doi: 10.1038/86723. [DOI] [PubMed] [Google Scholar]
- 21.Soukup GA, Breaker RR. Engineering precision RNA molecular switches. Proc Natl Acad Sci USA. 1999;96(7):3584–3589. doi: 10.1073/pnas.96.7.3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tang J, Breaker RR. Rational design of allosteric ribozymes. Chem biol. 1997;4(6):453–459. doi: 10.1016/s1074-5521(97)90197-6. [DOI] [PubMed] [Google Scholar]
- 23.Tang J, Breaker RR. Mechanism for allosteric inhibition of an ATP-sensitive ribozyme. Nucleic Acids Res. 1998;26(18):4214–4221. doi: 10.1093/nar/26.18.4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Y, Breaker RR. Deoxyribozymes: new players in the ancient game of biocatalysis. Curr Opin Struct Biol. 1999;9(3):315–323. doi: 10.1016/S0959-440X(99)80042-6. [DOI] [PubMed] [Google Scholar]
- 25.Wang DY, Lai BHY, Sen D. A general strategy for effector-mediated control of RNA-cleaving ribozymes and DNA enzymes. J Mole Biol. 2002;318(1):33–43. doi: 10.1016/S0022-2836(02)00046-3. [DOI] [PubMed] [Google Scholar]
- 26.Levy M, Ellington AD. ATP-dependent allosteric DNA enzymes. Chem Biol. 2002;9(4):417–426. doi: 10.1016/s1074-5521(02)00123-0. [DOI] [PubMed] [Google Scholar]
- 27.Robertson MP, Ellington A. In vitro selection of an allosteric ribozyme that transduces analytes to amplicons. Nat Biotechnol. 1999;17(1):62–66. doi: 10.1038/5236. [DOI] [PubMed] [Google Scholar]
- 28.Tang Z, Mallikaratchy P, Yang R. et al. Aptamer switch probe based on intramolecular displacement. J Am Chem Soc. 2008;130(34):11268–11269. doi: 10.1021/ja804119s. [DOI] [PubMed] [Google Scholar]
- 29.Zhen SJ, Chen LQ, Xiao SJ. et al. Carbon nanotubes as a low background signal platform for a molecular aptamer beacon on the basis of long-range resonance energy transfer. Anal Chem. 2010;82(20):8432–8437. doi: 10.1021/ac100709s. [DOI] [PubMed] [Google Scholar]
- 30.Du Y, Li B, Wang E. “Fitting” makes “sensing” simple: label-free detection strategies based on nucleic acid aptamers. Acc Chem Res. 2012;46(2):203–213. doi: 10.1021/ar300011g. [DOI] [PubMed] [Google Scholar]
- 31.Seemann IT, Singh V, Azarkh M. et al. Small-molecule-triggered manipulation of DNA three-way junctions. J Am Chem Soc. 2011;133(13):4706–4709. doi: 10.1021/ja110986u. [DOI] [PubMed] [Google Scholar]
- 32.He JL, Wu ZS, Zhou H. et al. Fluorescence aptameric sensor for strand displacement amplification detection of cocaine. Anal Chem. 2010;82(4):1358–1364. doi: 10.1021/ac902416u. [DOI] [PubMed] [Google Scholar]
- 33.Zhou R, Xu C, Dong J. et al. Labeling-free fluorescent detection of DNA hybridization through FRET from pyrene excimer to DNA intercalator SYBR green I. Biosens Bioelectron. 2015;65:103–107. doi: 10.1016/j.bios.2014.10.029. [DOI] [PubMed] [Google Scholar]
- 34.Lin Y, Tao Y, Ren J. et al. Highly sensitive and selective detection of thiol-containing biomolecules using DNA-templated silver deposition. Biosens Bioelectron. 2011;28(1):339–343. doi: 10.1016/j.bios.2011.07.040. [DOI] [PubMed] [Google Scholar]
- 35.Liu X, Shi L, Hua X. et al. Target-induced conjunction of Ssplit aptamer fragments and assembly with a Wwater-soluble conjugated polymer for improved protein detection. ACS Appl Mater interfaces. 2014;6(5):3406–3412. doi: 10.1021/am405550j. [DOI] [PubMed] [Google Scholar]
- 36.Lv F, Liu L, Wang S. Signal amplifying optical DNA detection on solid support with fluorescent conjugated polymers. Curr Org Chem. 2011;15(4):548–556. [Google Scholar]
- 37.Chen SJ, Huang CC, Chang HT. Enrichment and fluorescence enhancement of adenosine using aptamer-gold nanoparticles, PDGF aptamer, and Oligreen. Talanta. 2010;81(1):493–498. doi: 10.1016/j.talanta.2009.12.030. [DOI] [PubMed] [Google Scholar]
- 38.Zhu Y, Hu X, Shi S. et al. Ultrasensitive and universal fluorescent aptasensor for the detection of biomolecules (ATP, adenosine and thrombin) based on DNA/Ag nanoclusters fluorescence light-up system. Biosens Bioelectron. 2016;79:205–212. doi: 10.1016/j.bios.2015.12.015. [DOI] [PubMed] [Google Scholar]
- 39.Zhou Z, Du Y, Dong S. DNA-Ag nanoclusters as fluorescence probe for turn-on aptamer sensor of small molecules. Biosens Bioelectron. 2011;28(1):33–37. doi: 10.1016/j.bios.2011.06.028. [DOI] [PubMed] [Google Scholar]
- 40.Jiang Y, Fang X, Bai C. Signaling aptamer/protein binding by a molecular light switch complex. Anal Chem. 2004;76(17):5230–5235. doi: 10.1021/ac049565u. [DOI] [PubMed] [Google Scholar]
- 41.Li B, Wei H, Dong S. Sensitive detection of protein by an aptamer-based label-free fluorescing molecular switch. Chem Commun. 2006;(1):73–75. doi: 10.1039/b612080f. [DOI] [PubMed] [Google Scholar]
- 42.Wang J, Jiang Y, Zhou C. et al. Aptamer-based ATP assay using a luminescent light switching complex. Anal Chem. 2005;77(11):3542–3546. doi: 10.1021/ac050165w. [DOI] [PubMed] [Google Scholar]
- 43.Deng W, Xie F, Baltar HT. et al. Metal-enhanced fluorescence in the life sciences: here, now and beyond. Phys Chem Chem Phys. 2013;15:15695–15708. doi: 10.1039/c3cp50206f. [DOI] [PubMed] [Google Scholar]
- 44.Wang K, Liao J, Yang X. et al. A non-labelled aptasensor for highly sensitive detection of ATP and thrombin based on metal-enhanced PicoGreen fluorescence. Biosens Bioelectron. 2015;63:172–177. doi: 10.1016/j.bios.2014.07.022. [DOI] [PubMed] [Google Scholar]
- 45.Song Q, Peng M, He D. et al. A fluorescent aptasensor for amplified label-free detection of adenosine triphosphate based on core-shell Ag@ SiO2 nanoparticles. Biosens Bioelectron. 2016;77:237–241. doi: 10.1016/j.bios.2015.09.008. [DOI] [PubMed] [Google Scholar]
- 46.Wang Y, Liu B. ATP detection using a non-labelled DNA aptamer and a cationic tetrahedralfluorene. Analyst. 2008;133:1593–1598. doi: 10.1039/b806908e. [DOI] [PubMed] [Google Scholar]
- 47.Li Y, Li Y, Wang X. et al. A non-labelled conjugated polymer-based fluorescence assay for the determination of adenosine triphosphate and alkaline phosphatase. New J Chem. 2014;38:4574–4579. [Google Scholar]
- 48.Rupcich N, Nutiu R, Li Y. et al. Entrapment of fluorescent signaling DNA aptamers in sol-gel-derived silica. Anal Chem. 2005;77(14):4300–4307. doi: 10.1021/ac0506480. [DOI] [PubMed] [Google Scholar]
- 49.Carrasquilla C, Li Y, Brennan JD. Surface immobilization of structure-switching DNA aptamers on macroporous sol- gel-derived films for solid-phase biosensing applications. Anal Chem. 2011;83(3):957–965. doi: 10.1021/ac102679r. [DOI] [PubMed] [Google Scholar]
- 50.Hui CY, Li Y, Brennan JD. Fluorescence analysis of the properties of structure-switching DNA aptamers entrapped in sol-gel-derived silica materials. Chem Mater. 2014;26(5):1896–1904. [Google Scholar]
- 51.Nutiu R, Li Y. Aptamers with fluorescence-signaling properties. Methods. 2005;37:16–25. doi: 10.1016/j.ymeth.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 52.Zeng X, Zhang X, Yang W. et al. Fluorescence detection of adenosine triphosphate through an aptamer-molecular beacon multiple probe. Anal Biochem. 2012;424:8–11. doi: 10.1016/j.ab.2012.01.021. [DOI] [PubMed] [Google Scholar]
- 53.Zhao M, Liao L, Wu M. et al. Double-receptor sandwich supramolecule sensing method for the determination of ATP based on uranyl-salophen complex and aptamer. Biosens Bioelectron. 2012;34:106–111. doi: 10.1016/j.bios.2012.01.025. [DOI] [PubMed] [Google Scholar]
- 54.Song W, Zhang Q, Xie X. et al. Fluorescence aptameric sensor for isothermal circular strand-displacement polymerization amplification detection of adenosine triphosphate. Biosens Bioelectron. 2014;61:51–56. doi: 10.1016/j.bios.2014.04.030. [DOI] [PubMed] [Google Scholar]
- 55.Cho EJ, Yang L, Levy M. et al. Using a deoxyribozyme ligase and rolling circle amplification to detect a non-nucleic acid analyte, ATP. J Am Chem Soc. 2005;127(7):2022–2023. doi: 10.1021/ja043490u. [DOI] [PubMed] [Google Scholar]
- 56.Liu M, Song J, Shuang S. et al. A graphene-based biosensing platform based on the release of DNA probes and rolling circle amplification. ACS Nano. 2014;8(6):5564–5573. doi: 10.1021/nn5007418. [DOI] [PubMed] [Google Scholar]
- 57.Garai-Ibabe G, Möller M, Saa L. et al. Peroxidase-mimicking DNAzyme modulated growth of CdS nanocrystalline structures in situ through redox Rreaction: Application to development of genosensors and aptasensors. Anal Chem. 2014;86(20):10059–64. doi: 10.1021/ac502360y. [DOI] [PubMed] [Google Scholar]
- 58.Guo Y, Liu J, Yang G. et al. Multiple turnovers of DNAzyme for amplified detection of ATP and reduced thiol in cell homogenates. Chem Commun. 2015;51(5):862–865. doi: 10.1039/c4cc08428d. [DOI] [PubMed] [Google Scholar]
- 59.Sun N, Deng C, Liu Y. et al. Optimization of influencing factors of nucleic acid adsorption onto silica-coated magnetic particles: Application to viral nucleic acid extraction from serum. J Chromatogr A. 2014;1325:31–39. doi: 10.1016/j.chroma.2013.11.059. [DOI] [PubMed] [Google Scholar]
- 60.Smith JE, Wang L, Tan W. Bioconjugated silica-coated nanoparticles for bioseparation and bioanalysis. TrAC Trends in Anal Chem. 2006;25(9):848–855. [Google Scholar]
- 61.Wang Y, Wang Y, Liu B. Fluorescent detection of ATP based on signaling DNA aptamer attached silica nanoparticles. Nanotechnology. 2008;19(41):415605–415611. doi: 10.1088/0957-4484/19/41/415605. [DOI] [PubMed] [Google Scholar]
- 62.He X, Li Z, Jia X. et al. A highly selective sandwich-type FRET assay for ATP detection based on silica coated photon upconverting nanoparticles and split aptamer. Talanta. 2013;111:105–110. doi: 10.1016/j.talanta.2013.02.050. [DOI] [PubMed] [Google Scholar]
- 63.Chen Z, Li G, Zhang L. et al. A new method for the detection of ATP using a quantum-dot-tagged aptamer. Anal Bioanal Chem. 2008;392(6):1185–118. doi: 10.1007/s00216-008-2342-z. [DOI] [PubMed] [Google Scholar]
- 64.Cai L, Chen ZZ, Dong XM. et al. Silica nanoparticles based label-free aptamer hybridization for ATP detection using hoechst33258 as the signal reporter. Biosens Bioelectron. 2011;29(1):46–52. doi: 10.1016/j.bios.2011.07.064. [DOI] [PubMed] [Google Scholar]
- 65.He Y, Tian J, Hu K. et al. An ultrasensitive quantum dots fluorescent polarization immunoassay based on the antibody modified Au nanoparticles amplifying for the detection of adenosine triphosphate. Anal Chim Acta. 2013;802:67–73. doi: 10.1016/j.aca.2013.10.011. [DOI] [PubMed] [Google Scholar]
- 66.Tang X, Bansaruntip S, Nakayama N. et al. Carbon nanotube DNA sensor and sensing mechanism. Nano Lett. 2006;6(8):1632–1636. doi: 10.1021/nl060613v. [DOI] [PubMed] [Google Scholar]
- 67.Zheng M, Jagota A, Semke ED. et al. DNA-assisted dispersion and separation of carbon nanotubes. Nat Mater. 2003;2(5):338–342. doi: 10.1038/nmat877. [DOI] [PubMed] [Google Scholar]
- 68.Yang R, Jin J, Chen Y. et al. Carbon nanotube-quenched fluorescent oligonucleotides: probes that fluoresce upon hybridization. J Am Chem Soc. 2008;130(26):8351–8358. doi: 10.1021/ja800604z. [DOI] [PubMed] [Google Scholar]
- 69.Boul PJ, Cho DG, Rahman GA. et al. Sapphyrin-nanotube assemblies. J Am Chem Soc. 2007;129(17):5683–5687. doi: 10.1021/ja069266h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nakayama-Ratchford N, Bangsaruntip S, Sun X. et al. Noncovalent functionalization of carbon nanotubes by fluorescein-polyethylene glycol: supramolecular conjugates with pH-dependent absorbance and fluorescence. J Am Chem Soc. 2007;129(9):2448–2449. doi: 10.1021/ja068684j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang L, Wei H, Li J. et al. A carbon nanotubes based ATP apta-sensing platform and its application in cellular assay. Biosens Bioelectron. 2010;25(8):1897–1901. doi: 10.1016/j.bios.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 72.Wang L, Zhu J, Han L. et al. Graphene-based aptamer logic gates and their application to multiplex detection. ACS Nano. 2012;6(8):6659–6666. doi: 10.1021/nn300997f. [DOI] [PubMed] [Google Scholar]
- 73.Liu J, Wang C, Jiang Y. et al. Graphene signal amplification for sensitive and real-time fluorescence anisotropy detection of small molecules. Anal Chem. 2013;85(3):1424–1430. doi: 10.1021/ac3023982. [DOI] [PubMed] [Google Scholar]
- 74.Liu Z, Chen S, Liu B. et al. Intracellular detection of ATP using an aptamer beacon covalently linked to graphene oxide resisting nonspecific probe displacement. Anal Chem. 2014;86(24):12229–12235. doi: 10.1021/ac503358m. [DOI] [PubMed] [Google Scholar]
- 75.Lu CH, Wang F, Willner I. Amplified optical aptasensors through the endonuclease-stimulated regeneration of the analyte. Chem Sci. 2012;3(8):2616–2622. [Google Scholar]
- 76.Zhou Z, Du Y, Dong S. Double-strand DNA-templated formation of copper nanoparticles as fluorescent probe for label-free aptamer sensor. Anal Chem. 2011;83(13):5122–5127. doi: 10.1021/ac200120g. [DOI] [PubMed] [Google Scholar]
- 77.Liu JM, Yan XP. Competitive aptamer bioassay for selective detection of adenosine triphosphate based on metal-paired molecular conformational switch and fluorescent gold nanoclusters. Biosens Bioelectron. 2012;36(1):135–141. doi: 10.1016/j.bios.2012.04.015. [DOI] [PubMed] [Google Scholar]
- 78.Xu Y, Xu J, Xiang Y. et al. Target-induced structure switching of hairpin aptamers for label-free and sensitive fluorescent detection of ATP via exonuclease-catalyzed target recycling amplification. Biosens Bioelectron. 2014;51:293–296. doi: 10.1016/j.bios.2013.08.002. [DOI] [PubMed] [Google Scholar]
- 79.Wei Y, Chen Y, Li H. et al. An exonuclease I-based label-free fluorometric aptasensor for adenosine triphosphate (ATP) detection with a wide concentration range. Biosens Bioelectron. 2015;63:311–316. doi: 10.1016/j.bios.2014.07.064. [DOI] [PubMed] [Google Scholar]
- 80.Li F, Du Z, Yang L. et al. Selective and sensitive turn-on detection of adenosine triphosphate and thrombin based on bifunctional fluorescent oligonucleotide probe. Biosens Bioelectron. 2013;41:907–910. doi: 10.1016/j.bios.2012.10.007. [DOI] [PubMed] [Google Scholar]
- 81.Zhang Z, Sharon E, Freeman R. et al. Fluorescence detection of DNA, adenosine-5′-triphosphate (ATP), and telomerase activity by zinc (II)-protoporphyrin IX/G-quadruplex labels. Anal Chem. 2012;84(11):4789–4797. doi: 10.1021/ac300348v. [DOI] [PubMed] [Google Scholar]
- 82.Yang B, Zhang XB, Kang LP. et al. Target-triggered cyclic assembly of DNA-protein hybrid nanowires for dual-amplified fluorescence anisotropy assay of small molecules. Anal Chem. 2013;85(23):11518–11523. doi: 10.1021/ac402781g. [DOI] [PubMed] [Google Scholar]
- 83.Tan Y, Zhang X, Xie Y. et al. Label-free fluorescent assays based on aptamer-target recognition. Analyst. 2012;137(10):2309–2312. doi: 10.1039/c2an16092g. [DOI] [PubMed] [Google Scholar]
- 84.Leung KH, Lu L, Wang M. et al. A label-free luminescent switch-on assay for ATP using a G-quadruplex-selective iridium (III) complex. PloS one. 2013;8(10):e77021. doi: 10.1371/journal.pone.0077021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lu L, Qian Y, Wang L. et al. Metal-enhanced fluorescence-based core-shell Ag@ SiO2 nanoflares for affinity biosensing via target-induced structure switching of aptamer. ACS Appl Mater Interfaces. 2014;6(3):1944–1950. doi: 10.1021/am4049942. [DOI] [PubMed] [Google Scholar]
- 86.Zhang Y, Li B, Yan C. et al. One-pot fluorescence detection of multiple analytes in homogenous solution based on noncovalent assembly of single-walled carbon nanotubes and aptamers. Biosens Bioelectron. 2011;26(8):3505–3510. doi: 10.1016/j.bios.2011.01.035. [DOI] [PubMed] [Google Scholar]
- 87.Cheng S, Zheng B, Wang M. et al. A target-triggered strand displacement reaction cycle: The design and application in adenosine triphosphate sensing. Anal Biochem. 2014;446:69–75. doi: 10.1016/j.ab.2013.10.021. [DOI] [PubMed] [Google Scholar]
- 88.Li Z, Wang Y, Liu Y. et al. A novel aptasensor for the ultra-sensitive detection of adenosine triphosphate via aptamer/quantum dot based resonance energy transfer. Analyst. 2013;138(17):4732–4736. doi: 10.1039/c3an00449j. [DOI] [PubMed] [Google Scholar]
- 89.Pu WD, Zhang L, Huang CZ. Graphene oxide as a nano-platform for ATP detection based on aptamer chemistry. Anal Meth. 2012;4(6):1662–1666. [Google Scholar]
- 90.Jin F, Lian Y, Li J. et al. Molecule-binding dependent assembly of split aptamer and γ-cyclodextrin: A sensitive excimer signaling approach for aptamer biosensors. Anal Chim Acta. 2013;799:44–50. doi: 10.1016/j.aca.2013.08.012. [DOI] [PubMed] [Google Scholar]
- 91.Sun N, Guo Q, Shao J. et al. A signal-on fluorescence biosensor for detection of adenosine triphosphate based on click chemistry. Anal Meth. 2014;6(10):3370–3374. [Google Scholar]
- 92.Park JW, Park Y, Kim BH. Quencher-free molecular aptamer beacons (QF-MABs) for detection of ATP. Bioorg Med Chem Lett. 2015;25(20):4597–600. doi: 10.1016/j.bmcl.2015.08.052. [DOI] [PubMed] [Google Scholar]
- 93.Li X, Wang Y, Luo J. et al. Sensitive detection of adenosine triphosphate by exonuclease III-assisted cyclic amplification coupled with surface plasmon resonance enhanced fluorescence based on nanopore. Sens Actuat B Chem. 2016;228:509–514. [Google Scholar]
- 94.Zhao Q, Lv Q, Wang H. Aptamer fluorescence anisotropy sensors for adenosine triphosphate by comprehensive screening tetramethylrhodamine labeled nucleotides. Biosens Bioelectron. 2015;70:188–193. doi: 10.1016/j.bios.2015.03.031. [DOI] [PubMed] [Google Scholar]
- 95.Li X, Peng Y, Chai Y. et al. A target responsive aptamer machine for label-free and sensitive non-enzymatic recycling amplification detection of ATP. Chem Commun. 2016;52(18):3673–3676. doi: 10.1039/c6cc00110f. [DOI] [PubMed] [Google Scholar]
- 96.Zheng D, Seferos DS, Giljohann DA. et al. Aptamer nano-flares for molecular detection in living cells. Nano lett. 2009;9(9):3258–3261. doi: 10.1021/nl901517b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Jin F, Zheng J, Liu C. et al. Dual-stimuli responsive i-motif/nanoflares for sensing ATP in lysosomes. Analyst. 2014;139:3714–3717. doi: 10.1039/c4an00153b. [DOI] [PubMed] [Google Scholar]
- 98.Wang W, Chen C, Li X. et al. A bioresponsive controlled-release bioassay based on aptamer-gated Au nanocages and its application in living cells. Chem Commun. 2015;51:9109–9112. doi: 10.1039/c5cc02452h. [DOI] [PubMed] [Google Scholar]
- 99.Qiang W. Hu H, Sun L, et al. Aptamer/polydopamine nanospheres nanocomplex for in situ molecular sensing in living cells. Anal Chem. 2015;87:12190–12196. doi: 10.1021/acs.analchem.5b03075. [DOI] [PubMed] [Google Scholar]
- 100.Wang Y, Li Z, Hu D. et al. Aptamer/graphene oxide nanocomplex for in situ molecular probing in living cells. J Am Chem Soc. 2010;132(27):9274–9276. doi: 10.1021/ja103169v. [DOI] [PubMed] [Google Scholar]
- 101.Wang Y, Li Z, Weber TJ. et al. In situ live cell sensing of multiple nucleotides exploiting DNA/RNA aptamers and graphene oxide nanosheets. Anal Chem. 2013;85(14):6775–6782. doi: 10.1021/ac400858g. [DOI] [PubMed] [Google Scholar]
- 102.Shrivastava S, Sohn IY, Son YM. et al. Real-time label-free quantitative fluorescence microscopy-based detection of ATP using a tunable fluorescent nano-aptasensor platform. Nanoscale. 2015;7:19663–19672. doi: 10.1039/c5nr05839b. [DOI] [PubMed] [Google Scholar]
- 103.Tan X, Chen T, Xiong X. et al. Semi-quantification of ATP in live cells using nonspecific desorption of DNA from graphene oxide as the internal reference. Anal Chem. 2012;84:8622–8627. doi: 10.1021/ac301657f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Jia L, Ding L, Tian J. et al. Aptamer loaded MoS2 nanoplates as nanoprobes for detection of intracellular ATP and controllable photodynamic therapy. Nanoscale. 2015;7:15953–15961. doi: 10.1039/c5nr02224j. [DOI] [PubMed] [Google Scholar]
- 105.Roy S, Soh JH, Gao Z. A microfluidic-assisted microarray for ultrasensitive detection of miRNA under an optical microscope. Lab Chip. 2011;11(11):1886–1894. doi: 10.1039/c0lc00638f. [DOI] [PubMed] [Google Scholar]
- 106.Zu Y, Ting AL, Gao Z. Visualizing low-level point mutations: Enzyme-like selectivity offered by nanoparticle probes. Small. 2011;7(3):306–310. doi: 10.1002/smll.201001774. [DOI] [PubMed] [Google Scholar]
- 107.Mirkin CA, Letsinger RL, Mucic RC. et al. A DNA-based method for rationally assembling nanoparticles into macroscopic materials. Nature. 1996;382(6592):607–609. doi: 10.1038/382607a0. [DOI] [PubMed] [Google Scholar]
- 108.Huo Y, Qi L, Lv XJ. et al. A sensitive aptasensor for colorimetric detection of adenosine triphosphate based on the protective effect of ATP-aptamer complexes on unmodified gold nanoparticles. Biosens Bioelectron. 2016;78:315–320. doi: 10.1016/j.bios.2015.11.043. [DOI] [PubMed] [Google Scholar]
- 109.Wang J, Lu J, Su S. et al. Binding-induced collapse of DNA nano-assembly for naked-eye detection of ATP with plasmonic gold nanoparticles. Biosens Bioelectron. 2015;65:171–175. doi: 10.1016/j.bios.2014.10.031. [DOI] [PubMed] [Google Scholar]
- 110.Li J, Fu HE, Wu LJ. et al. General colorimetric detection of proteins and small molecules based on cyclic enzymatic signal amplification and hairpin aptamer probe. Anal Chem. 2012;84:5309–5315. doi: 10.1021/ac3006186. [DOI] [PubMed] [Google Scholar]
- 111.Zhao W, Chiuman W, Brook MA. et al. Simple and rapid colorimetric biosensors based on DNA aptamer and noncrosslinking gold nanoparticle aggregation. ChemBioChem. 2007;78(7):727–731. doi: 10.1002/cbic.200700014. [DOI] [PubMed] [Google Scholar]
- 112.Li H, Rothberg L. Colorimetric detection of DNA sequences based on electrostatic interactions with unmodified gold nanoparticles. Proc Nat Acad Sci USA. 2004;101(39):14036–14039. doi: 10.1073/pnas.0406115101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wang J, Wang L, Liu X. et al. A gold nanoparticle-based aptamer target binding readout for ATP assay. Adv Mater. 2007;19(22):3943–3946. [Google Scholar]
- 114.Chiorcea-Paquim AM, Oliveira-Brett AM. Redox behaviour of G-quadruplexes. Electrochim Acta. 2014;126:162–170. [Google Scholar]
- 115.Li S, Wang L, Hao Y. et al. An ultrasensitive colorimetric aptasensor for ATP based on peptide/Au nanocomposites and hemin-G-quadruplex DNAzyme. RSC Adv. 2014;4(44):23185–23190. [Google Scholar]
- 116.Gong X, Li J, Zhou W. et al. Target recycling amplification for label-free and sensitive colorimetric detection of adenosine triphosphate based on un-modified aptamers and DNAzymes. Anal Chim Acta. 2014;828:80–84. doi: 10.1016/j.aca.2014.04.028. [DOI] [PubMed] [Google Scholar]
- 117.Elbaz J, Moshe M, Shlyahovsky B. et al. Cooperative multicomponent self-assembly of nucleic acid structures for the activation of DNAzyme cascades: A paradigm for DNA sensors and aptasensors. Chem Eur J. 2009;15(14):3411–3418. doi: 10.1002/chem.200802004. [DOI] [PubMed] [Google Scholar]
- 118.Garcia-Campana AM, Baeyens W. Chemiluminescence in Analytical Chemistry. Marcel Dekker, Inc, New York; 2001. [Google Scholar]
- 119.Huang X, Li L, Qian H. et al. A resonance energy transfer between chemiluminescent donors and luminescent quantum-dots as acceptors (CRET) Angew Chem Int Ed. 2006;118(31):5264–5267. doi: 10.1002/anie.200601196. [DOI] [PubMed] [Google Scholar]
- 120.Zhang S, Yan Y, Bi S. Design of molecular beacons as signaling probes for adenosine triphosphate detection in cancer cells based on chemiluminescence resonance energy transfer. Anal Chem. 2009;81(21):8695–8701. doi: 10.1021/ac901759g. [DOI] [PubMed] [Google Scholar]
- 121.Zhou ZM, Yu Y, Zhao YD. A new strategy for the detection of adenosine triphosphate by aptamer/quantum dot biosensor based on chemiluminescence resonance energy transfer. Analyst. 2012;137(18):4262–4266. doi: 10.1039/c2an35520e. [DOI] [PubMed] [Google Scholar]
- 122.Mun H, Jo EJ, Li T. et al. Homogeneous assay of target molecules based on chemiluminescence resonance energy transfer (CRET) using DNAzyme-linked aptamers. Biosens Bioelectron. 2014;58:308–313. doi: 10.1016/j.bios.2014.02.008. [DOI] [PubMed] [Google Scholar]
- 123.Freeman R, Liu X, Willner I. Chemiluminescent and chemiluminescence resonance energy transfer (CRET) detection of DNA, metal ions, and aptamer-substrate complexes using hemin/G-quadruplexes and CdSe/ZnS quantum dots. J Am Chem Soc. 2011;133(30):11597–11604. doi: 10.1021/ja202639m. [DOI] [PubMed] [Google Scholar]
- 124.Li Y, Ji X, Song W. et al. Design of a sensitive aptasensor based on magnetic microbeads-assisted strand displacement amplification and target recycling. Anal Chim Acta. 2013;770:147–152. doi: 10.1016/j.aca.2013.01.044. [DOI] [PubMed] [Google Scholar]
- 125.Hutter E, Fendler JH. Exploitation of localized surface plasmon resonance. Adv Mater. 2004;16(19):1685–1706. [Google Scholar]
- 126.Liu Q, Jing C, Zheng X. et al. Nanoplasmonic detection of adenosine triphosphate by aptamer regulated self-catalytic growth of single gold nanoparticles. Chem Commun. 2012;48(77):9574–9576. doi: 10.1039/c2cc34632j. [DOI] [PubMed] [Google Scholar]
- 127.Cheng S, Zheng B, Wang M. et al. Double-functionalized gold nanoparticles with split aptamer for the detection of adenosine triphosphate. Talanta. 2013;115:506–511. doi: 10.1016/j.talanta.2013.05.065. [DOI] [PubMed] [Google Scholar]
- 128.Piliarik M, Homola J. Surface plasmon resonance (SPR) sensors: approaching their limits? Opt Express. 2009;17(19):16505–16517. doi: 10.1364/OE.17.016505. [DOI] [PubMed] [Google Scholar]
- 129.Campion A, Kambhampati P. Surface-enhanced Raman scattering. Chem Soc Rev. 1998;27(4):241–250. [Google Scholar]
- 130.Xu H, Aizpurua J, Käll M. et al. Electromagnetic contributions to single-molecule sensitivity in surface-enhanced Raman scattering. Phys Rev E. 2000;62(3):4318. doi: 10.1103/physreve.62.4318. [DOI] [PubMed] [Google Scholar]
- 131.Fleischmann M, Hendra PJ, McQuillan AJ. Raman spectra of pyridine adsorbed at a silver electrode. Chem Phys Lett. 1974;26(2):163–166. [Google Scholar]
- 132.Nie S, Emory SR. Probing single molecules and single nanoparticles by surface-enhanced Raman scattering. Science. 1997;275(5303):1102–1106. doi: 10.1126/science.275.5303.1102. [DOI] [PubMed] [Google Scholar]
- 133.Pieczonka NP, Aroca RF. Single molecule analysis by surfaced-enhanced Raman scattering. Chem Soc Rev. 2008;37(5):946–954. doi: 10.1039/b709739p. [DOI] [PubMed] [Google Scholar]
- 134.Ye S, Xiao J, Guo Y. et al. Aptamer based SERS assay of ATP and lysozyme by using primer SelfGeneration. Chem Eur J. 2013;19(25):8111–8116. doi: 10.1002/chem.201300126. [DOI] [PubMed] [Google Scholar]
- 135.Jiang Y, Meng F, Qi D. et al. Gold nanotip array for ultrasensitive electrochemical sensing and spectroscopic monitoring. Small. 2013;9(13):2260–2265. doi: 10.1002/smll.201202620. [DOI] [PubMed] [Google Scholar]
- 136.Lu W, Band BS, Yu Y. et al. Resonance light scattering and derived techniques in analytical chemistry: past, present, and future. Microchim Acta. 2007;158(1-2):29–58. [Google Scholar]
- 137.Enriquez AC, Espejel IR, García EA. et al. Enhanced resonance light scattering properties of gold nanoparticles due to cooperative binding. Anal Bioanal Chem. 2008;391(3):807–815. doi: 10.1007/s00216-008-1829-y. [DOI] [PubMed] [Google Scholar]
- 138.Yue Q, Shen T, Wang J. et al. A reusable biosensor for detecting mercury (II) at the subpicomolar level based on “turn-on” resonance light scattering. Chem Commun. 2013;49(17):1750–1752. doi: 10.1039/c3cc38488h. [DOI] [PubMed] [Google Scholar]
- 139.Chen Z, Song T, Wang S. et al. Screening DNA-targeted anticancer drug in vitro based on the drug-conjugated DNA by resonance light scattering technique. Biosens Bioelectron. 2010;25(8):1947–1952. doi: 10.1016/j.bios.2010.01.011. [DOI] [PubMed] [Google Scholar]
- 140.Jiang Z, Huang Y, Liang A. et al. Resonance scattering detection of trace microalbumin using immunonanogold probe as the catalyst of Fehling reagent-glucose reaction. Biosens Bioelectron. 2009;24(6):1674–1678. doi: 10.1016/j.bios.2008.08.031. [DOI] [PubMed] [Google Scholar]
- 141.Liang A, Ouyang H, Jiang Z. Resonance scattering spectral detection of trace ATP based on label-free aptamer reaction and nanogold catalysis. Analyst. 2011;136(21):4514–4519. doi: 10.1039/c1an15542c. [DOI] [PubMed] [Google Scholar]
- 142.He Y, Liao L, Xu C. et al. Determination of ATP by resonance light scattering using a binuclear uranyl complex and aptamer modified gold nanoparticles as optical probes. Microchim Acta. 2015;182(1-2):419–426. [Google Scholar]
- 143.Sessler JL, Melfi PJ, Pantos GD. Uranium complexes of multidentate N-donor ligands. Coord Chem Rev. 2006;250(7):816–843. [Google Scholar]
- 144.Rudkevich DM, Verboom W, Brzozka Z. et al. Functionalized UO2 salenes: neutral receptors for anions. J Am Chem Soc. 1994;116(10):4341–4351. [Google Scholar]
- 145.Yang X, Gao Z. Enzyme-catalysed deposition of ultrathin silver shells on gold nanorods: a universal and highly efficient signal amplification strategy for translating immunoassay into a litmus-type test. Chem Commun. 2015;51(32):6928–6931. doi: 10.1039/c5cc01286d. [DOI] [PubMed] [Google Scholar]
- 146.Nutiu R, Li Y. Structure-switching signaling aptamers. J Am Chem Soc. 2003;125(16):4771–4778. doi: 10.1021/ja028962o. [DOI] [PubMed] [Google Scholar]
- 147.Sazani PL, Larralde R, Szostak JW. A small aptamer with strong and specific recognition of the triphosphate of ATP. J Am Chem Soc. 2004;126(27):8370–8371. doi: 10.1021/ja049171k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Yu P, He X, Zhang L. et al. Dual recognition unit strategy improves the specificity of the adenosine triphosphate (ATP) aptamer biosensor for cerebral ATP assay. Anal Chem. 2014;87(2):1373–1380. doi: 10.1021/ac504249k. [DOI] [PubMed] [Google Scholar]