Abstract
Background
Genotype frequencies for chemotherapy-induced nausea and vomiting (CINV)-related polymorphisms have not yet been reported for Japanese subjects.
Methods
We analyzed 10 genotype frequencies for following polymorphisms associated with the development of CINV: serotonin 5-HT3 receptors (HTR3); neurokinin-1 receptors (Tachykinin-1 receptors, TACR1); dopamine D2 receptors (DRD2); and catechol-O-methyltransferase (COMT).
Results
All polymorphisms were successfully genotyped in 200 Japanese subjects and were in Hardy-Weinberg equilibrium. Almost all genotype frequencies were similar to those in the HapMap database or in the previous reports, while frequencies for the Y192H polymorphism in TACR1 were different in Japanese subjects from those in a previous report.
Conclusions
The present study revealed genotype frequencies for polymorphisms, which were related to the appearance of CINV in Japanese subjects. Individual therapy based on genotype variations for each race is needed to allow cancer patients to undergo chemotherapy more safely and to understand etiology of CINV.
Keywords: Chemotherapy-induced nausea and vomiting, Gene polymorphism, Japanese population
Background
Chemotherapy-induced nausea and vomiting (CINV) is a common severe side effect for cancer patients undergoing emetic chemotherapy [1, 2]. CINV is a significant problem because it affects not only the quality of life (QOL) of the patient but also determines the possibility of chemotherapy continuation. Thus, it is extremely important to overcome CINV.
The involvement of 5-hydroxytryptamine (5-HT; serotonin) has been reported as a mechanism of CINV, which is released from enterochromaffin cells in the mucosa of the small intestine adjacent to vagal afferent neurons in response to the stimulation of anti-cancer drugs [3, 4]. The released 5-HT activates serotonin 5-HT3 receptors of the medulla via the area postrema and the medial solitary nucleus, ultimately leading to a severe emetic response [5, 6]. Therefore, serotonin 5-HT3 receptor antagonists can significantly improve CINV [7]. According to the American Society of Clinical Oncology guidelines, an emetic prophylaxis for high-emetogenic-risk chemotherapy should include a serotonin 5-HT3 receptor antagonist, dexamethasone, and aprepitant [8], which, in combination, provides the best antiemetic efficacy [9].
Serotonin 5-HT3 receptors are members of the superfamily of Cys-loop ligand-gated ion channels [10]. There are five subunits encoded by different genes in the human genome: serotonin 5-HT3A, 5-HT3B, 5-HT3C, 5-HT3D, and 5-HT3E receptors [11–13]. Serotonin 5-HT3A and 5-HT3B receptors are expressed in the hippocampus, spleen, kidney, small intestine, and colon [14]. Serotonin 5-HT3A receptors are mainly involved in the formation of functional receptors [12], and it is the only subunit capable of forming functional homopentameric receptors [15]. The other subunits form functional receptors only when their receptor is co-expressed with serotonin 5-HT3A receptors [16–18].
CINV can also occur via other physiological neurotransmitters, including substance P [19], neurokinin-1 (NK-1) [20, 21], dopamine [21], and catechol-O-methyltransferase (COMT) [22]. Substance P is a member of the neurokinin family of peptides, which includes NK-1. NK-1 receptors (Tachykinin-1 receptors; TACR1) are located in the gut, the area postrema, and the nucleus tractus solitaries [4]. NK-1 exerts its biological effects by acting in the vomiting center in central NK-1 receptors. Thus, NK-1 receptor antagonists have recently been recommended when starting chemotherapy. The COMT enzyme modulates neurotransmission by metabolizing dopamine, which is known to play a role in the development of nausea and vomiting. Dopamine D2 receptor blockade in the area postrema and the vomiting center has an antiemetic effect. Thus, it is expected that polymorphisms of the COMT would have effects on dopamine-related pathogenesis, treatment, and adverse events [22].
Despite improvements in antiemetic treatment with serotonin 5-HT3 receptor antagonists, a considerable number of patients still suffer from CINV. Of patients that received an emetic prophylaxis, 20–38 % and 50–60 % showed delayed nausea and vomiting reactions, respectively [2, 23]. One potential reason for this effect is due to individual genetic differences in the function of their receptors and enzymes. Polymorphisms in their genes could serve as a predictive factor for CINV in patients undergoing moderately emetogenic chemotherapy [24], although there have been no reports confirming this genetic variation in a Japanese cancer patients. In this study, therefore, we analyzed genotype frequencies for polymorphisms of the HTR3, TACR1, DAD2, and COMT genes in Japanese subjects, which are associated with the development of CINV.
Methods
Study population
Japanese subjects, who presented at the Preventive Health Service Department of Nagoya University Hospital to have a physical checkup were recruited for this study. The sample of 200 subjects was recruited randomly from unrelated healthy individuals (average age: 49.9 years; range: 25–89 years) of which 125 were male and 75 were female, under institutionally approved internal review board protocols, with informed consent. This study was approved by the ethics committee of Nagoya University Graduate School of Medicine. This study was also performed according to Good Clinical Practice guidelines. The written informed consent documents were obtained from all subjects.
DNA isolation
Genomic DNA was extracted from peripheral blood using QIAamp® DNA Blood Mini Kit (QIAGEN; Valencia, CA, USA) following the manufacturer’s spin protocol instructions. Purified genomic DNA adjusted to a concentration of 10 ng/μL was stored at −30 °C until analysis.
Target gene polymorphisms
We analyzed the following 10 gene polymorphisms, which are known to be closely related to CINV (Fig. 1).
Serotonin 5-HT3 receptor genes: HTR3A (GenBank accession no. DQ050460) and HTR3B (DQ050462) in chromosome 11 (11q23.1), and HTR3C (AF459285) in chromosome 3 (3q27). We analyzed the untranslated region of HTR3A (rs1062613) [25], the exon regions of HTR3B (rs1176744 and Ala223Thr) [25, 26], the promoter region of HTR3B (−100_-102AAG deletion) [25], and the exon region of HTR3C (rs6766410) [24].
NK-1 receptor genes: TACR1 (AY420417) in chromosome 2 (2p13.1-p12). We analyzed the exon regions of TACR1 (Y192H and rs17838409).
Dopamine D2 receptor genes: DRD2 (AY418851) in chromosome 11 (11q22-23). We analyzed the exon region (rs6277) and the intron region (rs1076560).
COMT gene: COMT (DQ040245) in chromosome 22. We analyzed the exon region (rs4680) [22].
Fig. 1.
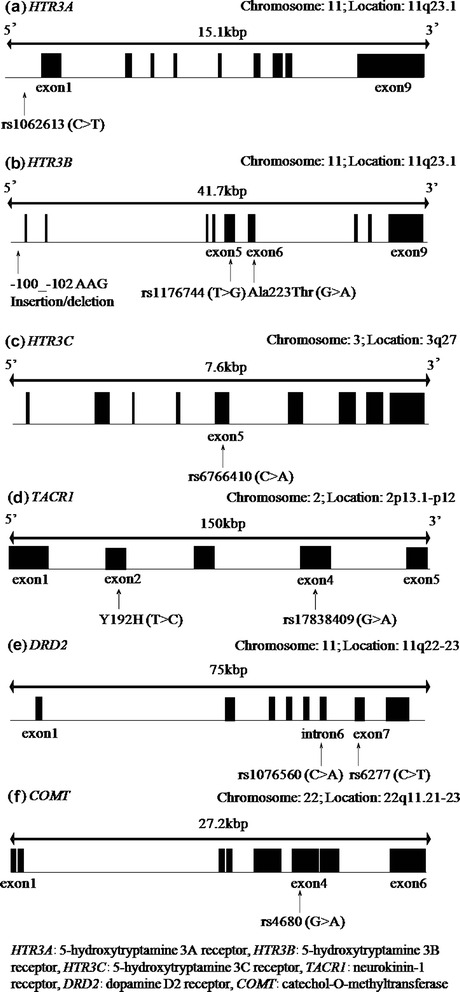
Location of gene polymorphisms. Representative gene structure, location of exons, and polymorphisms in (a) HTR3A serotonin 5-HT3A receptors, (b) HTR3B serotonin 5-HT3B receptors, (c) HTR3C serotonin 5-HT3C receptors, (d) TACR1 neurokinin-1 receptors, (e) DRD2 dopamine D2 receptors, and (f) COMT catechol-O-methyltransferase. For all polymorphisms, the accession numbers of the dbSNP database are indicated
Genotyping
Determination of allele variations among subjects was performed using the TaqMan® allelic discrimination assay (TaqMan® 5′-exonuclease allelic discrimination assay; Applied Biosystems; Foster City, CA, USA). The polymerase chain reaction (PCR) mixture contained 1 μL of genomic DNA, fluorescence probes (TaqMan® 20×, 40×, and 80× probes; Applied Biosystems), 2× TaqMan® Universal PCR Master Mix (Applied Biosystems), and distilled water (Wako; Tokyo, Japan) up to a total 10 μL volume in a 96-well microplate. Gene fragments were amplified by PCR using the Applied Biosystems Real-time PCR System. Amplification conditions were as follows: initial denaturation for 10 min at 95 °C, followed by 50 cycles of denaturation at 92 °C for 15 s, and annealing and extension at 58 °C for 1 min.
Deletion mutation screening
Deletion mutation screening was conducted by direct sequencing analysis. PCRs comprised 1 μL of DNA in the presence of 10× Buffer (TaKaRa; Shiga, Japan), dNTP (2.5 mM dNTPmix; TaKaRa), 20 μM forward/reverse primer (AAG deletion R/F; Rikaken; Nagoya, Japan), and rTagDNA polymerase (TaKaRa). The PCR mixture was amplified with PCR Thermal Cycler Dice® Standard (TaKaRa). The conditions of amplification were 94 °C for 1 min; 40 cycles of 94 °C for 30 s, 55 °C for 30 s, and elongation at 74 °C for 1 min; 1 cycle at 72 °C for 7 min. PCR products were purified from the reaction mixture using distilled water. Cycle sequencing was performed using the Big Dye® Terminator v1, Cycle Sequence Kit (Applied Biosystems) up to 20 μL, and then cleaning step and loaded onto an ABI PRISM® 310 Genetic Analyzer sequencer (Applied Biosystems).
Statistical analysis
Hardy-Weinberg equilibrium testing was performed using Haploview 4.1 software.
Results
The genotype and allele frequencies of the polymorphisms are shown in Table 1.
Table 1.
Genotype distribution
| No. | Gene | Localization | SNP | Exchange | N | Genotype frequencies (%) | MAF | HWE | ||
|---|---|---|---|---|---|---|---|---|---|---|
| −/− | +/− | +/+ | ||||||||
| 1 | HTR3A | 5′ UTR | rs1062613 | C > T | 200 | 150 (75.0) | 48 (24.0) | 2 (1.0) | 0.13 | 0.64 |
| 2 | HTR3B | Promotor | −100_-102 AAG insertion/deletion | delAAG | 127 | 94 (74.0) | 27 (21.3) | 6 (4.7) | - | - |
| 3 | Exon 5 | rs1176744 | T > G | 198 | 102 (51.5) | 83 (41.9) | 13 (6.6) | 0.28 | 0.62 | |
| 4 | Exon 6 | Ala223Thr | G > A | 200 | 200 (100) | 0 (0) | 0 (0) | 0 | - | |
| 5 | HTR3C | Exon 5 | rs6766410 | A > C | 191 | 79 (41.4) | 81 (42.4) | 31 (16.2) | 0.37 | 0.80 |
| 6 | TACR1 | Exon 2 | Y192H | T > C | 200 | 200 (100) | 0 (0) | 0 (0) | 0 | - |
| 7 | Exon 4 | rs17838409 | G > A | 200 | 200 (100) | 0 (0) | 0 (0) | 0 | - | |
| 8 | DRD2 | Exon 7 | rs6277 | C > T | 200 | 179 (89.5) | 21 (10.5) | 0 (0) | 0.06 | 0.98 |
| 9 | Intron 6 | rs1076560 | C > A | 200 | 76 (38.0) | 95 (47.5) | 29 (14.5) | 0.38 | 1.00 | |
| 10 | COMT | Exon 4 | rs4680 | G > A | 197 | 88 (44.6) | 85 (43.2) | 24 (12.2) | 0.34 | 0.87 |
HWE Hardy-Weinberg equilibrium, MAF minor allele frequency, HTR3A serotonin 5-HT3A receptors, HTR3B serotonin 5-HT3B receptors, HTR3C serotonin 5-HT3C receptors, TACR1 tachykinin-1 receptors, DRD2 dopamine D2receptors, COMT catechol-O-methyltransferase
The major alleles
The major alleles were as follows: the C allele for HTR3A (rs1062613), the T and G alleles for HTR3B (rs1176744 and Ala223Thr), the A allele for HTR3C (rs6766410), the T and G alleles for TACR1 (Y192H and rs17838409), the C allele for DRD2 (rs6277 and rs1076560), and the G allele for COMT (rs4680).
Genotype frequencies of serotonin 5-HT3 receptors
The genotype frequencies of serotonin 5-HT3 receptors were as follows. For HTR3A (rs1062613), the C/C, C/T, and T/T genotype frequencies were 75.0 % (n = 150), 24.0 % (n = 48), and 1.0 % (n = 2), respectively. For HTR3B (rs1176744), the T/T, T/G, and G/G genotype frequencies were 51.5 % (n = 102), 41.9 % (n = 83), and 6.6 % (n = 13), respectively. For HTR3B (Ala223Thr), the G/G, G/A, and A/A genotype frequencies were 100 % (n = 200), 0 % (n = 0), and 0 % (n = 0), respectively. For HTR3C (rs6766410), the A/A, A/C, and C/C genotype frequencies were 41.4 % (n = 79), 42.4 % (n = 81), and 16.2 % (n = 31), respectively.
On the other hand, for HTR3B (−100_-102AAG deletion) (n = 127; 73 subjects not detected), the frequencies of insertion/insertion, insertion/deletion, and deletion/deletion mutations were 74.0 % (n = 94), 21.3 % (n = 27), and 4.7 % (n = 6), respectively.
Genotype frequencies of NK-1 receptors
The genotype frequencies of NK-1 receptors were follows. For TACR1 (Y192H), the T/T, T/C, and C/C genotype frequencies were 100 % (n = 200), 0 % (n = 0), and 0 % (n = 0), respectively. For TACR1 (rs1738409), the G/G, G/A, and A/A genotype frequencies were 100 % (n = 200), 0 % (n = 0), and 0 % (n = 0), respectively.
Genotype frequencies of dopamine D2 receptors and COMT
The dopamine-related genotype frequencies were follows. For DRD2 (rs6277), the C/C, C/T, and T/T genotype frequencies were 89.5 % (n = 179), 10.5 % (n = 21), and 0 % (n = 0), respectively. For DRD2 (rs1076560), the C/C, C/A, and A/A genotype frequencies were 38.0 % (n = 76), 47.5 % (n = 95), and 14.5 % (n = 29), respectively. For COMT (rs4680), the G/G, G/A, and A/A genotype frequencies were 44.6 % (n = 88), 43.2 % (n = 85), and 12.2 % (n = 24), respectively.
Minor allele frequencies (MAFs)
The MAFs were as follows: 0.13 for rs1062613 (n = 200), 0.28 for rs1176744 (n = 198; 2 subjects not detected), 0.37 for rs6766410 (n = 191; 9 subjects not detected), 0.06 for rs6277 (n = 200), 0.38 for rs1076560 (n = 200), 0.34 for rs4680 (n = 197; 3 subjects not detected), and 0 for the other polymorphisms. There was no deviation from Hardy-Weinberg equilibrium detected (P > 0.05).
Discussions
We investigated genotype frequencies for polymorphisms related to the appearance of CINV in a Japanese population. HTR3B (Ala223Thr) and DRD2 (rs1076560) were particularly first study in Asian. Our results suggest that determining the genotype of these polymorphisms except for TACR1 and HTR3B can help to inform individually based medication for treating or preventing CINV using genomic information for the Japanese cancer patients.
As a general rule, the genotype of a sample used in a genetic analysis must conform to Hardy-Weinberg equilibrium [27]. Sample size (200 subjects) was small to reach conclusive findings, whereas the results could be reliable in Hardy-Weinberg equilibrium, which reflects a population’s actual genetic structure over time with the genetic structure. Our results showed that the genotype frequencies for polymorphisms were similar to those in the HapMap database or reported previously [28], suggesting that they are reliable. Namely, the MAFs of HTR3A (rs1062613, n = 86, Asian), HTR3B (rs1176744, n = 90, Asian), HTR3C (rs6766410, n = 88, Asian), DRD2 (rs6277, n = 82, Asian), DRD2 (rs1076560, n = 98, European), COMT (rs4680, n = 88, Asian), and TACR1 (rs17838409, n = 226, Asian) were 0.151, 0.30, 0.36, 0.049, 0.12, 0.24, and 0.004, respectively. Although HTR3B (Ala223Thr) is not indicated in the HapMap database (http://www.1000genomes.org/), the MAF has been reported to be 0.002 in Caucasians [26].
In a previous study, the HTR3 polymorphisms were shown to serve as a predictive factor for CINV [28]. Vomiting occurred in 50 % of patients with the C/C genotype of HTR3C (rs6766410), compared to only 19 % and 22 % in patients with the A/A and A/C genotypes, respectively [24]. These findings indicated that individual genetic differences affected the response to anti-emetic drugs. Patients with the 100_-102AAG deletion (deletion/deletion) showed vomiting more frequently than those with insertion/insertion and insertion/deletion mutations of this gene [25]. In the present Japanese sample, the −100_-102AAG deletion was not detected in 127 subjects. Although the reasons are unknown, the amount of DNA analyzed might have been too small to detect this deletion. Further investigations are needed to elucidate the role of the −100_-102AAG deletion in relation to CINV in Japanese subjects, including improving method of efficient DNA extraction.
Polymorphisms of the TACR1 (Y192H and rs17838409) are associated with the binding ability of substance P to NK-1 receptors [29], and have thus far been found only in the African-American population. It is therefore suggested that polymorphisms of the TACR1 are not clinically relevant for the Japanese population.
Gene polymorphisms have been associated with changing the expression level of gene or protein function. HTR3A (rs1062613), which is intronic polymorphism in the 5′ untranslated regions (5′UTRs) of HTR3A, affects the expression level of the downstream HTR3A [30]. Amino acid substitutions of HTR3B (rs1176744 and Ala223Thr) are related to receptor functional disorders [31]. Desensitization of serotonin 5-HT3 receptors does not occur in subjects with the HTR3B (rs1176744) polymorphism, which changes a tyrosine residue to a serine residue [32]. The T allele in DRD2 (rs6277) reduces the stabilization of the dopamine D2 receptors by changing the folded structure of the mRNA [33]. Thus, reduced dopamine D2 receptor (DRD2) binding was found to be associated with the C/C genotype of the rs6277 polymorphism of the DRD2 gene [34]. Subjects with an A allele in DRD2 (rs1076560) have a reduced ability to synthesize dopamine D2 receptors [35]. A missense variant of COMT (rs4680) also leads to an amino acid change. The Val version of COMT (G at rs4680) is associated with higher COMT enzyme activity leading to lower levels of dopamine in the brain, while the Met version (A at rs4680) is associated with lower enzyme activity and higher dopamine levels [22].
Determining an individual’s genotype is important to predict the clinical responses to chemotherapy, whereas it is difficult to incorporate the rapidly accumulating genome information for the Japanese population because of genetic differences among races. There are no studies the difference in Japanese and other races for the methods of preventing adverse effects related to CINV. As one of the individual-based medication for adverse effects induced by chemotherapy, UDP-glucuronosyltransferases (UGT) 1A1*28 has been suggested to be related to neutropenia induced by irinotecan, a topoisomerase inhibitor used in combination with other chemotherapeutic agents. FDA (Food and Drug Administration) recommends that patients with UGT1A1*28 are treated at the small doses of irinotecan, because the frequency of UGT1A1*28 in Caucasian is higher than that in Asian [36]. We believe that our results help to determine individual-based medications for treating and/or preventing CINV in Japanese cancer patients. Further studies are needed to confirm the relationship between gene polymorphisms and the efficacy of antiemetic therapies on CINV in Japanese cancer patients. These support the idea of establishing individualized supportive therapies (some additional prophylactic antiemetics) for CINV, and contribute to the development of more effective and safer chemotherapies.
Conclusions
We identified the genotype frequencies for polymorphisms related to the mechanism of appearance of CINV in Japanese subjects. Our study theoretically contributes to increasing the safety of chemotherapy with supportive therapy to prevent CINV and increase the QOL of cancer patients.
Abbreviations
5-HT, 5-hydroxytryptamine; ABCB1, ATP binding cassette subfamily B member 1; CINV, chemotherapy-induced nausea and vomiting; COMT, catechol-O-methyltransferase; DRD2, dopamine D2 receptors; HTR, serotonin 5-HT receptors; MAF, minor allele frequencies; NK-1, neurokinin-1; OPRM1, opioid receptor mu 1; PCR, polymerase chain reaction; QOL, quality of life; TACR1, tachykinin-1 receptors
Acknowledgments
This study was supported by The ‘Academic Frontier’ Project for Private Universities (2007–2011); Grants-in-Aid for Scientific Research C from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan [24590219, 16K08421], and The Adaptable and Seamless Technology Transfer Program Through Target-driven R&D, Japan Science and Technology Agency [AS251Z03018]. We would like to thank Ms. Chihiro Murosaki, Ms. Rina Kishi, and all staff members of Faculty of Pharmacy, Meijo University that were involved in this study.
Authors’ contributions
AG, HK, KY, TN, and YN designed the research. HK, MM, KI, YS, YN, and YH performed the research. AG and YS analyzed the data. AG, HK, YH, and YN wrote the article. All authors reviewed and edited the final report. All authors read and approved the final manuscript.
Competing interests
Yoshinori Hasegawa had received grant from Organization Astellas, Chugai and Eli Lilly, and speaker’s bureau for Organization Boehringer, MSD and Eli Lilly; the remaining authors has no competing interests.
Ethics approval and consent to participate
The present study was performed in accordance with the protocol approved by Ethics Committee of Nagoya University Hospital (approval number: 465–4).
References
- 1.Navari RM. Management of chemotherapy-induced nausea and vomiting: focus on newer agents and new uses for older agents. Drugs. 2013;73:249–62. doi: 10.1007/s40265-013-0019-1. [DOI] [PubMed] [Google Scholar]
- 2.Slatkin NE. Cannabinoids in the treatment of chemotherapy-induced nausea and vomiting: beyond prevention of acute emesis. J Support Oncol. 2007;5:1–9. [PubMed] [Google Scholar]
- 3.Hornby PJ. Central neurocircuitry associated with emesis. Am J Med. 2001;111(Suppl 8A):106S–112S. doi: 10.1016/S0002-9343(01)00849-X. [DOI] [PubMed] [Google Scholar]
- 4.Rojas C, Slusher BS. Pharmacological mechanisms of 5-HT3 and tachykinin NK1 receptor antagonism to prevent chemotherapy-induced nausea and vomiting. Eur J Pharmacol. 2012;684:1–7. doi: 10.1016/j.ejphar.2012.01.046. [DOI] [PubMed] [Google Scholar]
- 5.Tyers MB, Freeman AJ. Mechanism of the anti-emetic activity of 5-HT3 receptor antagonists. Oncology. 1992;49:263–8. doi: 10.1159/000227054. [DOI] [PubMed] [Google Scholar]
- 6.Minami M, Endo T, Hamaue N, Hirafuji M. Serotonin and anticancer drug-induced emesis. Yakugaku Zasshi. 2004;124:491–507. doi: 10.1248/yakushi.124.491. [DOI] [PubMed] [Google Scholar]
- 7.Rojas C, Raje M, Tsukamoto T, Slusher BS. Molecular mechanisms of 5-HT3 and NK1 receptor antagonists in prevention of emesis. Eur J Pharmacol. 2014;722:26–37. doi: 10.1016/j.ejphar.2013.08.049. [DOI] [PubMed] [Google Scholar]
- 8.Basch E, Prestrud AA, Hesketh PJ, Kris MG, Feyer PC, Somerfield MR, et al. Antiemetics: American Society of Clinical Oncology clinical practice guideline update. J Clin Oncol. 2011;29:4189–98. doi: 10.1200/JCO.2010.34.4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hesketh PJ, Grunberg SM, Gralla RJ, Warr DG, Roila F, de Wit R, Aprepitant Protocol 052 Study Group et al. The oral neurokinin-1 antagonist aprepitant for the prevention of chemotherapy-induced nausea and vomiting: a multinational, randomized, double-blind, placebo-controlled trial in patients receiving high-dose cisplatin--the Aprepitant Protocol 052 Study Group. J Clin Oncol. 2003;21:4112–9. doi: 10.1200/JCO.2003.01.095. [DOI] [PubMed] [Google Scholar]
- 10.Reeves DC, Lummis SC. The molecular basis of the structure and function of the 5-HT3 receptor: a model ligand-gated ion channel (review) Mol Membr Biol. 2002;19:11–26. doi: 10.1080/09687680110110048. [DOI] [PubMed] [Google Scholar]
- 11.Davies PA, Pistis M, Hanna MC, Peters JA, Lambert JJ, Hales TG, et al. The 5-HT3B subunit is a major determinant of serotonin-receptor function. Nature. 1999;397:359–363. doi: 10.1038/16941. [DOI] [PubMed] [Google Scholar]
- 12.Niesler B, Frank B, Kapeller J, Rappold GA. Cloning, physical mapping and expression analysis of the human 5-HT3 serotonin receptor-like genes HTR3C, HTR3D and HTR3E. Gene. 2003;310:101–111. doi: 10.1016/S0378-1119(03)00503-1. [DOI] [PubMed] [Google Scholar]
- 13.Thompson AJ, Lummis SC. 5-HT3 receptors. Curr Pharm Des. 2006;12:3615–30. doi: 10.2174/138161206778522029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yaakob N, Malone DT, Exintaris B, Irving HR. Heterogeneity amongst 5-HT3 receptor subunits: is this significant? Curr Mol Med. 2011;11:57–68. doi: 10.2174/156652411794474392. [DOI] [PubMed] [Google Scholar]
- 15.Thompson AJ, Lummis SC. The 5-HT3 receptor as a therapeutic target. Expert Opin Ther Targets. 2007;11:527–540. doi: 10.1517/14728222.11.4.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barrera NP, Herbert P, Henderson RM, Martin IL, Edwardson JM. Atomic force microscopy reveals the stoichiometry and subunit arrangement of 5-HT3 receptors. Proc Natl Acad Sci USA. 2005;102:12595–600. doi: 10.1073/pnas.0503253102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lummis SC. 5-HT3 receptors. J Biol Chem. 2012;287:40239–45. doi: 10.1074/jbc.R112.406496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Niesler B, Walstab J, Combrink S, Möller D, Kapeller J, Rietdorf J, et al. Characterization of the novel human serotonin receptor subunits 5-HT3C, 5-HT3D, and 5-HT3E. Mol Pharmacol. 2007;72:8–17. doi: 10.1124/mol.106.032144. [DOI] [PubMed] [Google Scholar]
- 19.Jordan K, Schmoll HJ, Aapro MS. Comparative activity of antiemetic drugs. Crit Rev Oncol Hematol. 2007;61:162–75. doi: 10.1016/j.critrevonc.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 20.Takahashi T, Nakamura Y, Tsuya A, Murakami H, Endo M, Yamamoto N. Pharmacokinetics of aprepitant and dexamethasone after administration of chemotherapeutic agents and effects of plasma substance P concentration on chemotherapy-induced nausea and vomiting in Japanese cancer patients. Cancer Chemother Pharmacol. 2011;68:653–9. doi: 10.1007/s00280-010-1519-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hesketh PJ. Chemotherapy-induced nausea and vomiting. N Engl J Med. 2008;358:2482–2494. doi: 10.1056/NEJMra0706547. [DOI] [PubMed] [Google Scholar]
- 22.Hosak L. Role of the COMT gene Val158Met polymorphism in mental disorders: a review. Eur Psychiatry. 2007;22:276–281. doi: 10.1016/j.eurpsy.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 23.Gregory RE, Ettinger DS. 5-HT3 receptor antagonists for the prevention of chemotherapy-induced nausea and vomiting. A comparison of their pharmacology and clinical efficacy. Drugs. 1998;55:173–189. doi: 10.2165/00003495-199855020-00002. [DOI] [PubMed] [Google Scholar]
- 24.Fasching PA, Kollmannsberger B, Strissel PL, Niesler B, Engel J, Kreis H, et al. Polymorphisms in the novel serotonin receptor subunit gene HTR3C show different risks for acute chemotherapy-induced vomiting after anthracycline chemotherapy. J Cancer Res Clin Oncol. 2008;134:1079–1086. doi: 10.1007/s00432-008-0387-1. [DOI] [PubMed] [Google Scholar]
- 25.Lehmann AS, Renbarger JL, McCormick CL, Topletz AR, Rouse C, Haas DM. Pharmacogenetic predictors of nausea and vomiting of pregnancy severity and response to antiemetic therapy: a pilot study. BMC Pregnancy Childbirth. 2013;13:132. doi: 10.1186/1471-2393-13-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tremblay PB, Kaiser R, Sezer O, Rosler N, Schelenz C, Possinger K, et al. Variations in the 5-hydroxytryptamine type3B receptor gene as predictors of the efficacy of antiemetic treatment in cancer patients. J Clin Oncol. 2003;21:2147–55. doi: 10.1200/JCO.2003.05.164. [DOI] [PubMed] [Google Scholar]
- 27.Smits KM, Schouten JS, Smits LJ, Stelma FF, Nelemans P, Prins MH. A review on the design and reporting of studies on drug-gene interaction. J Clin Epidemiol. 2005;58:651–654. doi: 10.1016/j.jclinepi.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 28.Pud D, Har-Zahav G, Laitman Y, Rubinek T, Yeheskel A, Ben-Ami S, et al. Association between variants of 5-hydroxytryptamine receptor 3C (HTR3C) and chemotherapy-induced symptoms in women receiving adjuvant treatment for breast cancer. Breast Cancer Res Treat. 2014;144:123–31. doi: 10.1007/s10549-014-2832-y. [DOI] [PubMed] [Google Scholar]
- 29.Randolph GP, Simon JS, Arreaza MG, Qiu P, Lachowicz JE, Duffy RA. Identification of single-nucleotide polymorphisms of the human neurokinin 1 receptor gene and pharmacological characterization of a Y192H variant. Pharmacogenomics J. 2004;4:394–402. doi: 10.1038/sj.tpj.6500276. [DOI] [PubMed] [Google Scholar]
- 30.Niesler B, Flohr T, Nothen MM, Fischer C, Rietschel M, Franzek E, et al. Association between the 5′ UTR variant C178T of the serotonin receptor gene HTR3A and bipolar affective disorder. Pharmacogenetics. 2001;11:471–5. doi: 10.1097/00008571-200108000-00002. [DOI] [PubMed] [Google Scholar]
- 31.Niesler B, Kapeller J, Hammer C, Rappold G. Serotonin type 3 receptor genes: HTR3A, B, C, D. E. Pharmacogenomics. 2008;9:501–4. doi: 10.2217/14622416.9.5.501. [DOI] [PubMed] [Google Scholar]
- 32.Walstab J, Hammer C, Bönisch H, Rappold G, Niesler G. Naturally occurring variants in the HTR3B gene significantly alter properties of human heteromeric 5-hydroxytryptamine-3A/B receptors. Pharmacogenet Genomics. 2008;18:793–802. doi: 10.1097/FPC.0b013e3283050117. [DOI] [PubMed] [Google Scholar]
- 33.Duan J, Wainwright MS, Comeron JM, Saitou N, Sanders AR, Gelernter J, et al. Synonymous mutations in the human dopamine receptor D2 (DRD2) affect mRNA stability and synthesis of the receptor. Hum Mol Genet. 2003;12:205–16. doi: 10.1093/hmg/ddg055. [DOI] [PubMed] [Google Scholar]
- 34.Hirvonen MM, Lumme V, Hirvonen J, Pesonen U, Någren K, Vahlberg T, et al. C957T polymorphism of the human dopamine D2 receptor gene predicts extrastriatal dopamine receptor availability in vivo. Prog Neuropsychopharmacol Biol Psychiatry. 2009;33:630–6. doi: 10.1016/j.pnpbp.2009.02.021. [DOI] [PubMed] [Google Scholar]
- 35.Bertolino A, Fazio L, Caforio G, Blasi G, Rampino A, Romano R, et al. Functional variants of the dopamine receptor D2 gene modulate prefronto-striatal phenotypes in schizophrenia. Brain. 2009;132:417–25. doi: 10.1093/brain/awn248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Innocenti F, Undevia SD, Iyer L, Chen PX, Das S, Kocherginsky M, et al. Genetic variants in the UDP-glucuronosyltransferase 1A1 gene predict the risk of severe neutropenia of irinotecan. J Clin Oncol. 2004;22:1382–8. doi: 10.1200/JCO.2004.07.173. [DOI] [PubMed] [Google Scholar]


