Fig. 3. Inhibition of KRASG12C requires GTPase activity and is attenuated by nucleotide exchange.
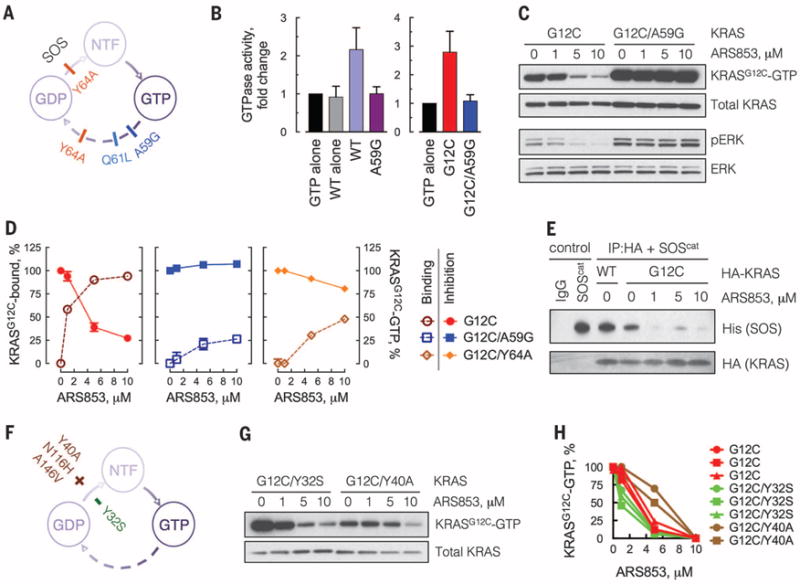
(A) Schematic of the mutations used to block the GTPase activity of KRAS. NTF, nucleotide free. (B) KRAS mutants, expressed and affinity purified from HEK293 cells, were subjected to a GTPase reaction. The orthophosphate product was measured by the Cytophos reagent and expressed as fold change over GTP alone (n = 3, mean and SEM). (C) HEK293 cells expressing HA-tagged KRAS constructs were treated with ARS853 for 5 hours. Cell extracts were evaluated by RBD:PD and immunoblotting with an anti-HA antibody, to determine the effect on GTP-bound mutant KRASG12C. A representative of three independent experiments is shown. (D) Extracts from cells expressing the KRAS constructs treated as shown were evaluated by mass spectrometry to determine the percentage of KRASG12C bound to the drug. KRASG12C-GTP levels in the same extracts were determined as in (C) and quantified by densitometry (n = 3, mean and SEM). (E) HEK293 cells expressing the constructs shown were treated with ARS853 for 5 hours. HA-tagged KRAS was immunoprecipitated (IP) and subjected to a binding assay with the catalytic domain of SOS (SOScat). (F) Schematic of the mutations used to biochemically increase (+) or decrease (−) the nucleotide exchange function of KRASG12C. (G) HEK293 cells expressing the indicated constructs were analyzed as in (C). (H) The level of active KRAS-GTP was quantified by densitometry. Each replicate is shown. A similar result was obtained in H358 cells (fig. S6F). The baseline level of KRASG12C/Y40A detected by the RBD:PD is low because the Y40 mutation impairs the interaction of RAS with RAF (33). See also the effects of N116H and A146V mutations, which increase nucleotide exchange without affecting RAS-RAF interaction (fig. S6G).
