FIGURE 1.
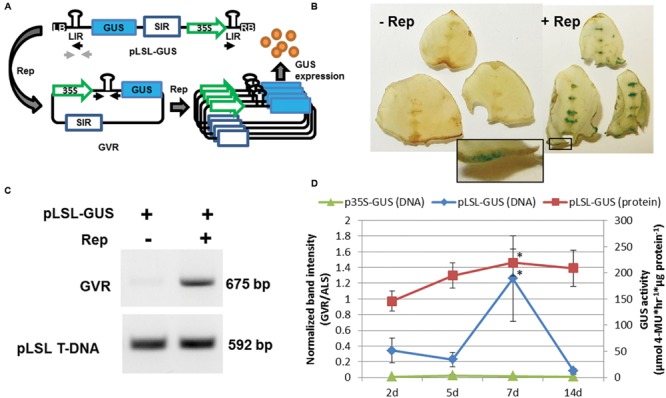
Delivery of the geminivirus replicon (GVR) to potato leaf explants. (A) Schematic of pLSL-GUS T-DNA used for Agrobacterium-mediated delivery of GVRs to potato leaf tissues. Replicase (Rep) is delivered on a separate p35S T-DNA binary vector (not shown). LB and RB; left and right T-DNA borders, respectively. SIR and LIR; short and long intergenic regions, respectively. 35S; cauliflower mosaic virus promoter. Blue rectangle; GUS coding sequence. Black and light gray arrows; priming sites used for PCR detection of circularized GVRs and pLSL T-DNA, respectively. (B) GUS staining of potato leaf explants transformed with pLSL-GUS. Potato leaf explants were transformed with pLSL-GUS in the presence (+Rep) or absence (-Rep) of Rep, and stained for GUS activity 7 days post-inoculation (dpi). Inset is magnification of wounded areas (open black rectangle). Images are from Désirée. (C) PCR detection of circularized GVRs in potato leaf explants transformed with pLSL-GUS. Leaf explants transformed with pLSL-GUS in the presence (+) or absence (-) of Rep were sampled for PCR detection of circularized GVRs (675 bp), and the pLSL T-DNA (592 bp) using priming sites from panel (A). Images are from Désirée. (D) Time-course of GVRs in potato leaf explants constitutively expressing Rep. Leaf explants prepared from a mutant potato line, D52 (Supplementary Figure S2) were transformed with pLSL-GUS and control p35S-GUS T-DNAs, and sampled after 2, 5, 7, and 14 dpi for quantitative end-point PCR of circularized GVRs (DNA; primary axis) and GUS activity quantification (protein; secondary axis). Error bars represent standard deviations from three biological replications. ∗P < 0.05; 2 dpi.
