Figure 1.
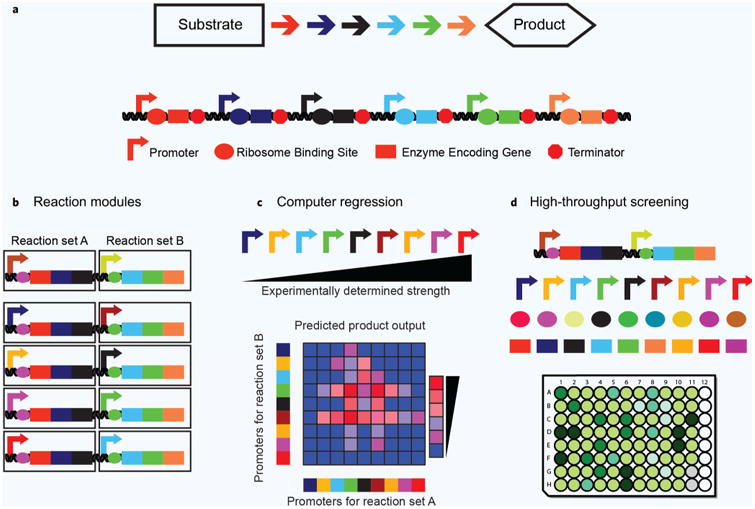
There are three main strategies to sample sequence-flux space in metabolic pathways. (a) A model pathway. Each arrow represents a different reaction chemistry used to convert an initial substrate into a value-added product. Expression elements like promoters and ribosome binding sites (RBS) facilitate expression of the enzyme-encoding gene sequences. (b) Individual reaction chemistries are grouped into modules. Expression of these modules is varied, reducing the combinatorial search space. (c) Given a training set of different sequences with a given output, models can be harnessed to predict optimal pathway expression levels. (d) Advanced DNA assembly methods can be used to create unique pathway variants that are then assessed using high-throughput screens or selections.
