Abstract
An estimated three billion people are at risk of Dengue virus (DENV) infection worldwide and there are currently no approved therapeutic interventions for DENV infection. Due to the relatively small size of the DENV genome, DENV is reliant on host factors throughout the viral life cycle. The inducible form of Heat Shock Protein 70 (Hsp70i) has been implicated as a host factor in DENV pathogenesis, however the complete role remains to be elucidated. Here we further illustrate the importance of Hsp70i in dengue virus pathogenesis and describe the antiviral activity of the allosteric small molecule inhibitor that is selective for Hsp70i, called HS-72. In monocytes, Hsp70i is expressed at low levels preceding DENV infection, but Hsp70i expression is induced upon DENV infection. Targeting Hsp70i with HS-72, results in a dose dependent reduction in DENV infected monocytes, while cell viability was maintained. HS-72 works to reduce DENV infection by inhibiting the entry stage of the viral life cycle, through disrupting the association of Hsp70i with the DENV receptor complex. This work highlights Hsp70i as an antiviral target and HS-72 as a potential anti-DENV therapeutic agent.
Keywords: Heat Shock Protein 70, Dengue Virus, Small Molecule Inhibitor, Antiviral Target, Antiviral Therapy
1. Introduction
Dengue virus (DENV) is the leading cause of arthropod transmitted disease in humans, consisting of four distinct serotypes (DENV 1–4), which are transmitted to humans through mosquito vectors, primarily Aedes aegypti and Aedes albopictus (1–3). In 1970, DENV epidemics had only been reported in nine countries, but by 2009, DENV was reported to be endemic in over 100 countries worldwide (1). An estimated three billion people worldwide are at risk for infection and recent reports estimate that the number of infections each year could be close to 400 million (1, 4). Due to the expanding geographical distribution of DENV and the frequency of epidemics, the WHO has classified DENV as a major international public health concern (3). Infection can manifest as the self-limited febrile illness known as dengue fever, or the potentially fatal dengue hemorrhagic fever (DHF) and dengue shock syndrome (DSS) (5). DENV infection results in at least 500,000 hospitalizations, while DHF and DSS are the leading cause of hospitalization and death of children in eight countries (6, 7). The potential for DHF and DSS are increased in a second DENV infection, due to antibody dependent enhancement (ADE). Antibodies are produced in response to DENV infection for that serotype, which provides protection against the infecting serotype and cross-protection for the other three serotypes. ADE occurs in a secondary infection when the cross-neutralizing antibodies fall below the limit of protection and enhance the secondary serotype infecting host cells through binding Fcγ receptors and entry through Fcγ receptor-mediated endocytosis (8, 9). Currently there are no approved therapies or vaccines for DENV and the need to protect against all four serotypes is a challenge faced when treating DENV, which when coupled with the lack of vector control in endemic countries, stresses the need to develop novel and effective DENV therapeutics (4, 6, 10). Furthermore, if a vaccine fails to efficiently protect against one of the serotypes there is an increased possibility of severe dengue disease due to ADE.
DENV is a member of the genus Flavivirus with a genome that encodes for 3 structural proteins, the Capsid (C), premembrane/membrane (prM), and envelope (E), and 7 non-structural proteins, NS1, NS2A, NS2B, NS3, NS4A, NS4B, NS5 (11). Due to the relatively small genome, host factors are utilized for efficient replication and propagation of DENV, and recent studies have aimed to identify potential host factors (12–14). Targeting host proteins for antiviral therapeutics provides an advantage over targeting viral proteins because host proteins are not as susceptible to mutations that result in the development of drug resistant strains, which can occur when targeting viral proteins (15). One such host factor that has recently been identified that is involved in DENV pathogenesis is the Heat Shock Protein 70 family of proteins (Hsp70), including the stress inducible Hsp70 (Hsp70i) and the constitutively active Hsp70 (Hsc70) (16–18).
The Hsp70’s are protein chaperones that have many cellular functions, which include the folding of nascent proteins, refolding of misfolded proteins, and protein transport between cellular compartments (19). These functions are driven by ATP hydrolysis in the N-terminal nucleotide-binding domain (NBD). Structurally Hsp70 family members are similar and contain 3 functional domains, a NBD in the N-terminal region, a substrate-binding domain in the C-terminal region, and a linker in the middle (20). The Hsp70 family of proteins are evolutionary conserved and consist of several family members. These include the stress inducible Hsp70i (also called Hsp72, Hsp70–1, and HspA1A/HspA1B) as well as the constitutively active form, Hsc70 (19). Hsp70i is present at low levels in unstressed cells; however, expression levels rapidly increase in response to cellular stresses such as heat shock or in response to certain viral infections (21–25). Previous studies have shown that siRNA knockdown of Hsp70 results in decreased DENV RNA copy numbers in supernatants and decreased intracellular DENV load (17). Furthermore, Hsp70 was identified through affinity chromatography to be part of the DENV receptor complex, however it is not fully elucidated which member of the Hsp70 family is part of the complex (18). Hsp70 family members as a part of the DENV receptor complex is supported by previous studies showing a reduction in DENV infected human and mosquito cells when pretreated with Hsp70 antibodies (18, 26). Additionally, it has been shown that there is an increase in the number of DENV infected cells following heat shock (16). The increase in infection correlates with an increase in Hsp70 present on the cell surface, but the different Hsp70 family members are not discriminated (16). Therefore, the precise role of the inducible Hsp70 family member remains to be fully elucidated.
This work describes the antiviral activity of a recently identified allosteric small molecule inhibitor that is selective for Hsp70i, called HS-72 (27). Additionally, the role of Hsp70i as a host factor in DENV pathogenesis is further elucidated. We show that the antiviral activity of HS-72 is through disrupting entry of DENV into host cells. Our data indicates that Hsp70i associates with the DENV receptor complex to mediate entry and this association is disrupted by HS-72. This work highlights Hsp70i as an antiviral target and HS-72 as a potential therapeutic agent.
2. Methods
2.1. Cell lines, antibodies, and virus preparation
U937+DC-SIGN (dendritic cell-specific intercellular adhesion molecule-3-grabbing non-integrin) cells were provided by Aravinda de Silva (University of North Carolina at Chapel Hill) and were maintained in RPMI-1640 media supplemented with 10% heat inactivated FBS (Sigma-Aldrich; St. Louis, MO), 1% L-glutamine, 50μM beta-mercaptoethanol, 1% non-essential amino acids, and 1× penicillin/streptomycin. Huh7 cells were provided by Mariano Garcia-Blanco (University of Texas Medical Branch) and were maintained in DMEM media supplemented with 10% FBS (Sigma-Aldrich) and 1× penicillin/streptomycin. African green monkey Vero cells (ATCC CCL-81) and Aedes albopictus C6/36 cells (ATCC CRL-1660) were obtained from the American Type Culture Collection. Vero cells were maintained in DMEM media supplemented with 10% FBS and 1× penicillin/streptomycin. C6/36 cells were maintained in MEM media supplemented with 10% FBS, 1% HEPES, 1% sodium pyruvate, 1% non-essential amino acids, and 1× penicillin/streptomycin. All cell lines were maintained at 37°C and 5% CO2, expect for C6/36, which was maintained at 28°C and 5% CO2.
Hsp70i, Hsp90, DC-SIGN, and GAPDH antibodies were purchased from Cell Signaling Technology (Danvers, MA). The DENV E protein antibody, 4G2, was generously provided by Mariano Garcia-Blanco (University of Texas Medical Branch).
Dengue Virus 2-NGC (DENV) stocks were prepared by incubating 500μL of DENV in 4.5mL serum free media on confluent C6/36 cells. After 1 hour incubation, virus media was replaced with complete growth media supplemented with 2% FBS and 5mM HEPES. Cells were incubated for 72 hours and cell media was collected and centrifuged at 4,000 rpm for five minutes. The supernatants were saved and stored at −80°C. Virus titer was quantified by foci forming assay.
2.2. DENV proteomic profiling
Huh7 cells were infected with DENV at a multiplicity of infection (MOI) of ten. Two hours before cell harvest, cell media was spiked with 440μCi of [35S]-methionine and [35S]-cysteine (Easy tag EXPRESS35S; Perkin Elmer; Waltham, MA). Cells were lysed in lysis buffer (0.1% Triton; 150mM NaCl; 60mM MgCl2; 25mM Tris-HCl, pH 7.5; 1mM DTT, 1μM Microcystin (Cayman Chemical; Ann Arbor, MI), and 1× protease inhibitor tablet (Roche; Mannheim, Germany)), clarified by centrifugation, and incubated with a γ-phosphate ATP-affinity resin. The resin was washed 3× with low salt wash buffer (50mM Tris, pH 7.5; 150mM NaCl; and 60mM Mg Cl2) and bound proteins were removed from the resin by boiling in 5× SDS running buffer. Samples were subjected to SDS-PAGE and analyzed by silver stain. The gel was then dried and analyzed by autoradiogram for seven days. Indicated bands were excised and identified by matrix-assisted laser desorption-ionization mass spectrometry (MALDI-MS) on an ABSCIEX (Framingham, MA) TOF/TOF 5800 mass spectrometer.
2.3. Western Blotting
SDS-PAGE was carried out using CriterionTM Cell system using pre-casted 4–20% or 4–15% CriterionTM Tris-HCl gels (BioRad; Hercules, CA). Gels were run using the PowerPac basic power supply (BioRad) and were transferred to nitrocellulose (Fisher Scientific; Waltham, MA). Nitrocellulose membranes were blocked with 5% dry non-fat milk in Tris-buffered saline (TBS) with 0.01% Tween-20 (TBS-T) for 1 hour at room temperature. Membranes were incubated with primary antibodies overnight at 4°C. Next day membranes were washed 3X in TBS-T, incubated for 1 hour at room temperature with secondary antibodies, and further washed 3X in TBS-T. ECL Plus Western blotting reagent (Pierce Biotechnology; Rockford, IL) was used to detect antibodies. Where indicated, bands were quantified using ImageJ software (28).
2.4. Flow Cytometry
U937+DC-SIGN cells were plated at 3 × 105 cells/well and treated with the indicated compounds in a volume of 100μL. Cells were then infected with DENV at a MOI = 1, where indicated. To inactivate DENV, an equivalent of a MOI = 1 of DENV was heated at 55°C for 30 minutes prior to addition to cells, where indicated. After a 1 hour incubation, virus media was removed and compound containing media was added. Compounds were maintained for duration of experiments. At the indicated time points post-infection, cells were incubated in Fixable Viability Dye eFluor® 780 (eBioscience; San Diego, CA) following the manufacture protocol. Cells were washed 2× in PBS + 0.5% BSA (Sigma-Aldrich) and fixed in 2% PFA. Where indicated, cells were permeablized with ice-cold methanol. Cells were incubated in primary antibody followed by the matching Alexa Flour® 488 (Invitrogen; Carlsbad, CA) secondary antibody. Samples were collected using the BD Facs Canto II (BD bioscience; Franklin Lakes, NJ). Data was analyzed using FlowJo version 10 software (FlowJo, LLC, Ashland, OR). Non-specific isotype control antibody (Thermo Scientific; Rockford, IL) was used as a control.
2.5. Foci Forming Assay
Virus culture supernatant from U937+DC-SIGN cells treated with HS-72 in a volume of 100μL, was added to Vero cells in 10-fold serial dilutions. Vero cells were incubated for 1 hour with virus media. After this incubation, 200μL of 1:1 tragacanth gum/2× EMEM overlay with 2% FBS was added to each well and incubated for 4 days. The overlay solution was removed and cells were fixed with 4% PFA in PBS, permeablized in 0.5% Triton X-100, and blocked in PBS + 0.1% tween and 1% normal donkey serum (Millipore; Temecula, CA). Viral foci were identified using the DENV E protein antibody 4G2 followed by incubation with Alexa Fluor® 488 secondary antibody. Foci were visualized using an AMG EVOS® FL Imaging System (Life Technologies; Carlsbad, CA).
2.6. Time of Addition Assay
U937+DC-SIGN cells were plated at 3 × 105 cells/well. At the indicated time points, cells were treated with HS-72 in a volume of 100μL. At time point zero, cells were then infected with DENV at a MOI = 1. After a 1 hour incubation, virus media was removed. HS-72 was maintained from the time of treatment for duration of the assay. Cells were fixed in 4% PFA, permeablized in PBS + 0.5% Triton X-100, and blocked in PBS + 1% normal donkey serum. Infected cells were identified using the DENV E protein antibody 4G2 followed by incubation with Alexa Fluor® 488 secondary antibody. DAPI was used to designate cells. Cells were imaged using the Cellomics Array Scan VTI system (Thermo Scientific). Percent infection and cell number were determined using vHCS Scan software version 5 (Thermo Scientific).
2.7. Attachment and Entry Assay
U937+DC-SIGN cells were plated at 3 × 105 cells/well and treated with HS-72, in a volume of 100μL, for 1 hour, cells were then cooled to 4°C, and D ENV was added at a MOI = 1. After 1 hour, RNA was extracted from cells, which corresponds to the attachment stage. Separately, cells were warmed to 37°C for 1 hour, and RNA were extracted, which corresponds to the entry stage of the life cycle. RNA was extracted using the Arum RNA kit (BioRad) and cDNA was generated using the iScript cDNA synthesis kit (BioRad) according to the manufactures instructions. cDNA along with primers and SYBR® Green Supermix (Biorad) were added according to the manufactures instructions for qPCR. qPCR was run at an initial 95°C for 5 minutes, then 40 cycles of 95°C for 15 seconds and 60°C for 30 seconds, on a CFX384 Touch Real-Time PCR Detection System (BioRad). The fold change of viral RNA vs. GAPDH, which served as the control, was determined. Primers are as follows: DENV-FWD 5′-AATATGCTGAAACGCGAGAGA-3′, DENV-REV 5′-GGGATTGTTAGGAAACGAAGG-3′, GAPDH-FWD 5′-GAGTCAACGGATTTGGTCGT-3′, and GAPDH-REV 5′-TTGATTTTGGAGGGATCTCG-3′.
2.8. Biotin labeling of surface proteins
U937+DC-SIGN cells were plated at 3 × 105 cells/well and treated with the HS-72 in a volume of 100μL. Cells were then infected with DENV at a MOI = 1, where indicated. At the indicated time point post-infection, cells were put on ice in a 4°C cold room to maintain proteins on the cell surface. Modification of a previously published protocol was used for biotin labeling (29). Briefly, cells were washed 3× with PBS2+(PBS + 1.5mM MgCl2, 0.2mM Ca Cl2) and incubated with Sulfo-NHS-SS-biotin (Thermo Scientific) 2× for 15 minutes. Free biotin was quenched by incubating 2× for 15 minutes in PBS2+ supplemented with 100mM glycine. Cells were lysed in RPIA buffer (10mM Tris, pH 7.4; 150mM NaCl; 1 mM EDTA; 0.1% SDS; 1% Triton; 1% sodium deoxycholate) supplemented with 1× protease inhibitor tablet (Roche). Protein concentration was determined and 50μg of protein is incubated with Avidin sepharose resin. The resin was washed with RIPA buffer, incubated with 2× Laemmli sample buffer (BioRad) to cleave the NHS-SS-biotin disulfide bond, and the eluted proteins were subjected to SDS-PAGE and western blot analysis.
2.9. Proximity Ligation Assay
U937+DC-SIGN cells were plated at 3 × 105 cells/well and treated with HS-72 in a volume of 100μL. Cells were then infected with DENV at a MOI = 1, where indicated, attached to poly-lysine treated cover slips (VWR), and fixed in 4% PFA. Cells were washed in PBS, incubated in wheat germ agglutinin conjugated to Alexa flour 488 (WGA-488, Life Technologies) to stain cell membranes, blocked in 5% normal goat serum (AbCam; Cambridge, MA), and incubated with primary antibodies. Proximity Ligation Assay (PLA) reagents were used according to the manufactures instructions (Sigma-Aldrich). Briefly, PLA probes were added to samples, followed by the addition of ligation solution to hybridize and link the probes in a circle. The ligated circle serves as the template for rolling circle amplification (RCA) through addition of polymerase along with fluorescently labeled oligonucleotides, which will hybridize with the RCA product and yield a fluorescent signal. The cover slips were then mounted with mounting medium containing DAPI to slides and imaged using a Zeiss Axio Imager widefield fluorescence microscope.
2.10. Compounds
HS-72 (27) and HS-10 (30) were synthesized as previously described and Ribavirin was purchased from Sigma-Aldrich.
2.11. Statistical analysis
All statistical analysis conducted using a t test on GraphPad Prisim4 (La Jolla, CA). Significance determined as p < 0.05.
3. Results
3.1. Proteomic analysis shows Hsp70i is a DENV host factor
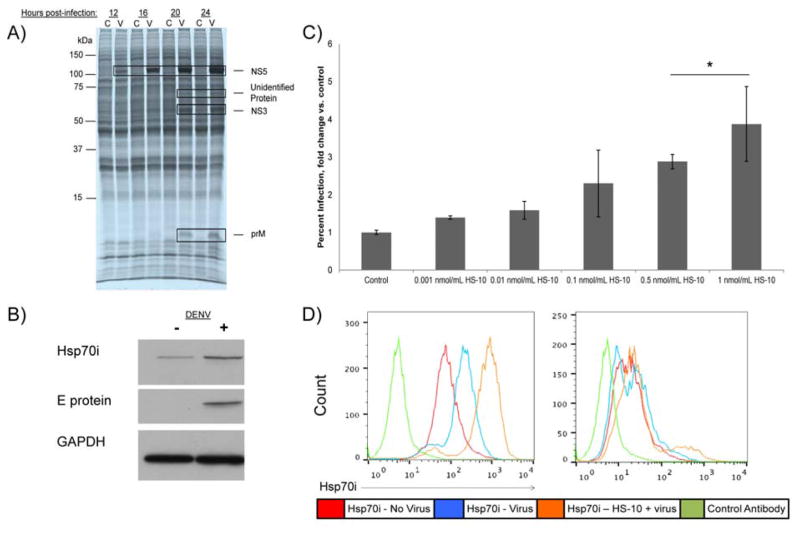
Previously, our laboratory has employed nucleotide affinity resins to capture and enrich en masse purine utilizing proteins (the purinome) from tumor cell and tissue extracts, with the goal of defining novel drug targets as well as to screen small molecule libraries for selective inhibitors (27, 31, 32). Purine-binding proteins, by virtue of their nucleotide binding pockets represent approximately 50% of the estimated druggable purinome and include a diverse range of enzyme classes e.g. protein kinases, non-protein kinases, heat shock proteins, metabolic enzymes, DNA and RNA binding proteins (33). Herein we employed an affinity resin based on ATP immobilized through its γ-phosphate to define host purine utilizing proteins that may be induced or activated in response to DENV infection (Fig. 1A). Huh7 cells were treated with [35S] methionine/cysteine and virus (MOI = 10) and infection followed over 24 hours. At the indicated time points the cells were stopped and the purinome isolated by passing the radio-labeled cellular homogenates over the ATP resin in parallel. Bound proteins were then characterized by SDS-PAGE, autoradiography and mass spectrometry (MS) (Fig. 1A). Fig. 1A shows marked increases in the translation of four ATP binding proteins in response to DENV-infection by 12–24 h post-infection at 105 kDa, 70 kDa, 65 kDa and 8 kDa. Three were identified by MS as virally encoded proteins NS5, NS3, and prM (Fig. 1A). Both NS5, and NS3 were expected to bind ATP via their respective polymerase, NTPase or helicase domains, however, the reasons for recovery of prM is unknown. All other proteins detected in the autoradiogram were unaffected by the infection, and identified by MS as host ATP binding proteins (Appendix S1). We suspected that the 70 kDa protein may be the inducible form of Hsp70, Hsp70i, based on previous reports by others describing induction of the protein following DENV infection (21–25). Western blot analysis with antibodies to Hsp70i confirmed this conclusion (Figs. 1B and S1A). To determine if the induction of Hsp70i was the result of a general cellular stress in response to DENV infection or likely to be facilitated by the virus itself, we induced expression of the protein in uninfected cells using the Hsp90 inhibitor, HS10 (30). Induction of Hsp70i expression is a signature response to all Hsp90 inhibitors targeting the ATP binding domain of the protein (34–36). Hsp90 exists in a complex with heat shock transcription factor 1 (HSF-1) in cells and upon Hsp90 inhibition, the complex is disassociated, which allows HSF-1 to translocate to the nucleus (37). After trimerization, HSF-1 binds to heat shock elements on heat shock gene promoters, which elicits an increase in Hsp70i transcription (37). The induction of Hsp70i by Hsp90 inhibition is thought to be a compensatory response to maintain cell viability (35, 37). Fig. S1B shows treating uninfected cells with HS-10 for 6 hours results in a dose dependent increase in Hsp70i expression. When we infected the Hsp70i induced cells with DENV we observed a correlation of infectivity with Hsp70i expression compared with non-HS10 treated cells (Figs. 1C and S1B–C). We also observed by flow cytometry an increase in surface Hsp70i expression, in both infected cells and infected cells pretreated with HS-10 (Figs. 1D and S1D). When HS-10 was administered without preincubation prior to addition of DENV, there is no effect on the percent of infected cells from non-treated cells (Fig. S1E). This finding is consistent with a requirement for the induction of Hsp70i expression, following Hsp90 inhibition, leading to increased infectivity. This experiment also showed that acute Hsp90 inhibition does not have acute antiviral activity against DENV in culture.
Fig. 1. Proteomic Analysis Shows Hsp70i is a DENV host factor.
(A) Labeling of Huh7 cells with [35S] methionine/cysteine prior to DENV infection reveals induction of 105 kDa, 70 kDa, 65 kDa, and 8 kDa proteins 12–24 hours post-infection. Huh7 cells were labeled with [35S] methionine/cysteine and infected with DENV, harvested at the indicated time points following infection, bound to a γ-phosphate ATP-sepharose resin, separated by SDS-PAGE, visualized by autoradiogram, and analyzed by mass spectrometry (MS). The DENV NS5, NS3, and prM proteins were identified as the 105 kDa, 65 kDa, and 8 kDa proteins, while the 70 kDa protein was not identified. Lanes labeled “C” indicate uninfected control samples; lanes labeled “V” indicated DENV infected samples. (B) Induction of Hsp70i was confirmed following DENV infection, in U937+DC-SIGN cells. Following 24 hours of DENV infection, samples were subjected to SDS-PAGE, and analyzed by Western blot. E protein serves as a positive control for infection, GAPDH serves as a loading control. (C) U937+DC-SIGN cells were pretreated with HS-10, to induce Hsp70i expression prior to DENV infection. This results in a dose dependent increase in infectivity, compared to cells that were not pretreated with HS-10. After HS-10 pretreatment for six hours, DENV was added to cells, and 24 hours post-infection cells were processed for flow cytometry to determine the percent infection. (Mean ± SEM. *, p<0.05 compared to control). (D) Total and Surface Hsp70i expression was induced in infected U937+DC-SIGN cells and infected cells pretreated with HS-10. The indicated samples were pretreated with HS-10, DENV was added to cells, and 24 hours post-infection cells were analyzed by flow cytometry for total and surface Hsp70i expression. Non-specific antibody used as control.
See also S1 Fig.
3.2. The Hsp70i inhibitor, HS-72, reduces DENV infection
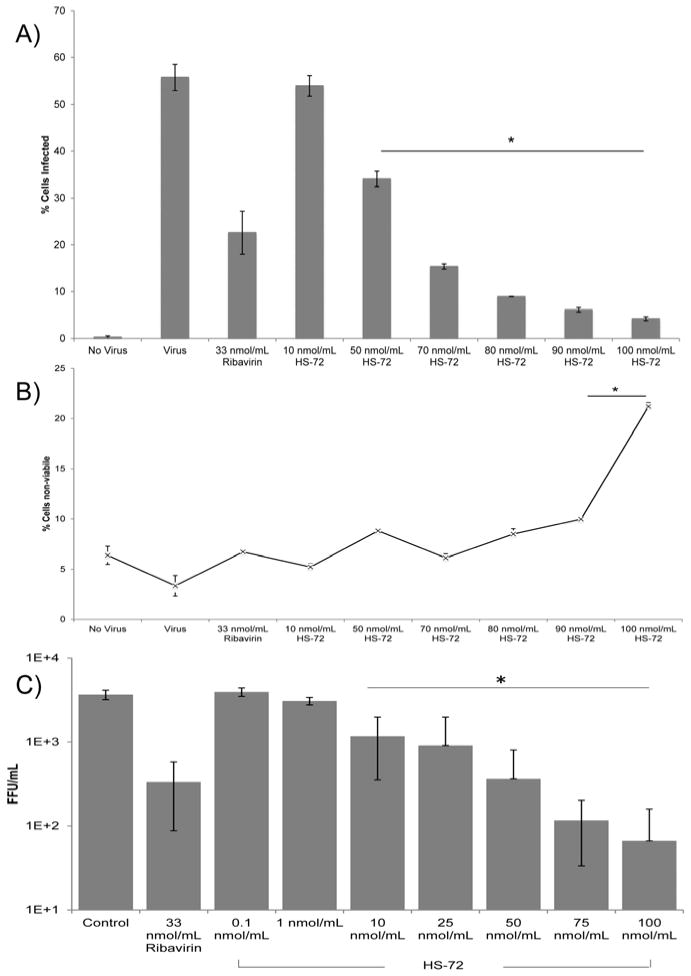
Recently we identified a highly specific allosteric small molecule inhibitor targeting Hsp70i, called HS-72 (27). The molecule discriminates Hsp70i from the closely related homolog, Hsc70, and exhibits bioavailability and efficacy in cell and animal models of human breast cancer (Howe et al. 2014). The molecule acts at an allosteric site on Hsp70i altering the affinity of the protein for ATP and thereby inhibiting its ATPase activity and chaperone functions. Given the effects of Hsp70i induction on increasing infectivity, we tested the antiviral activity of the molecule against DENV. The human monocytic cell line, U937, expressing the dendritic cell specific ICAM3-grabbing non-integrin (DC-SIGN) receptor, was treated with HS-72 or Ribavirin for 1 hour prior to addition of DENV. At 24 hours post-infection, the cells were analyzed by flow cytometry for infection and viability (Figs. 2A and 2B). Fig. 2 shows treatment with HS-72 results in a dose dependent reduction in DENV infected cells with no significant decrease in cell viability at <90 nmol/ml. These data indicate a targeted reduction in infection through Hsp70i inhibition as opposed to non-discriminant cell toxicity from HS-72 treatment. In addition to reducing DENV infected U937+DC-SIGN cells, treatment of Huh7 cells with HS-72 also results in a reduction in DENV infected cells (Fig. S2). This demonstrates the anti-DENV activity of HS-72 in both monocytes as well as liver cells, two distinct cell types that are susceptible to DENV infection. Furthermore, in foci forming assays, using the supernatants from U937+DC-SIGN cells to infect Vero cells, HS-72 shows a similar dose dependent decrease in DENV infection, with an EC50 of 22.8 nmol/ml (Fig. 2C).
Fig. 2. The Hsp70i inhibitor, HS-72, reduces DENV infection.
(A) The Hsp70i inhibitor, HS-72, yields a dose dependent reduction in DENV infection. U937+DC-SIGN cells were treated with HS-72 for 1 hour, DENV was added to cells, and 24 hours post-infection cells were processed for flow cytometry. DENV infection results in 56% of cells infected, with HS-72 reducing infection to 34% of cells infected at 50 nmol/ml, 15% at 70 nmol/ml, 9% at 80 nmol/ml, 6% at 90 nmol/ml, and 4% at 100 nmol/ml. Ribavirin was included as a control for reducing DENV infection, which reduced the percent of infected cells to 23% at 33 nmol/ml. Compounds were maintained on cells for the duration of the assay. An antibody for the DENV E protein coupled with a fluorescent secondary antibody was used to determine cells positive for DENV infection. (B) U937+DC-SIGN cells treated and infected as described in (A), were analyzed for cell viability by flow cytometry using an eFluor® 780 conjugated fixable viability dye. Cell viability was maintained when HS-72 <90 nmol/ml. This indicates a targeted reduction in DENV infection by HS-72, through maintaining cell viability. (C) HS-72 reduced DENV infection in a dose dependent manner as determined by a decrease in foci forming units, with a significant reduction compared to control at >10 nmol/ml. U937+DC-SIGN cells were treated with HS-72, following the treatment timeline as described in (A), after 24 hours supernatants were removed and added to Vero cells for a 1 hour incubation. A tragacanth gum solution was overlaid and cells were incubated for 72 hours. Next, cells were processed and probed with a DENV E protein antibody coupled with a fluorescent secondary antibody to determine infection. Foci were counted to determine the foci forming units/mL.
(A – C) Mean ± SEM. *, p<0.05 compared to control.
See also S2 Fig.
3.3. HS-72 inhibits DENV entry into host cells
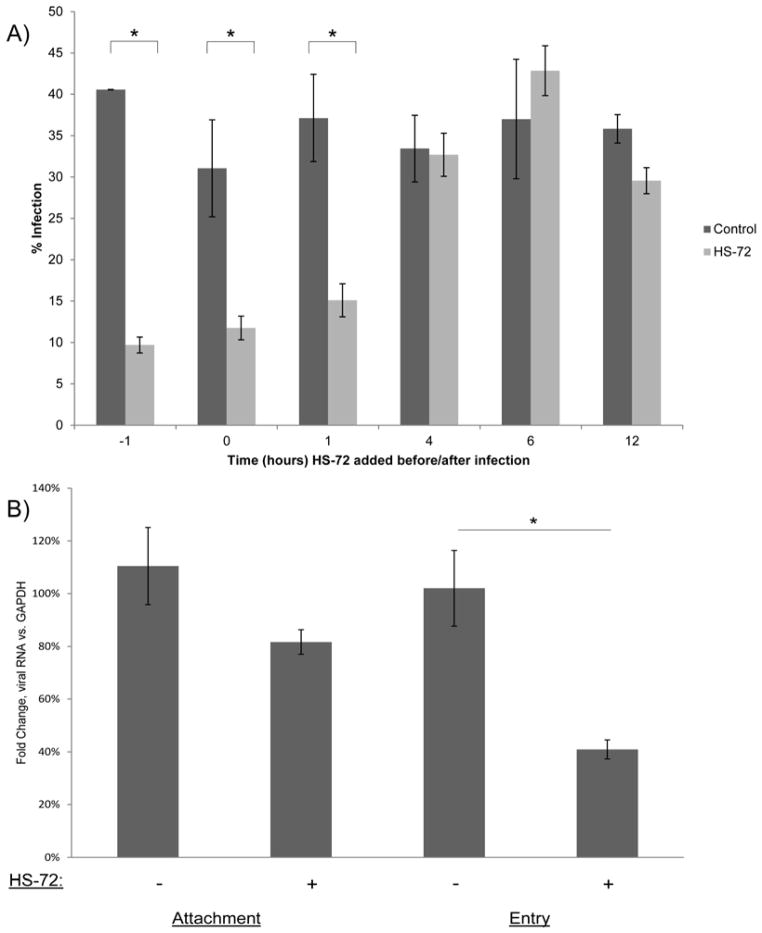
To determine the stage of the DENV life cycle inhibited by HS-72, U937+DC-SIGN cells were treated with the inhibitor 1 hour before addition of DENV, at the same time as DENV addition, and then 1 hour, 4 hours, 6 hours, and 12 hours post infection. At 24 hours after addition of DENV, percent infection and cell number were determined (Figs. 3A and S3). These studies showed that HS-72 was most effective when added prior to infection, or at the 0 or 60 minute time points, but ineffective when added 4 hours, 6 hours, or 12 hours post DENV infection (Fig. 3A). These data indicate HS-72 is most likely working to block the DENV life cycle at early phases. Importantly, as observed earlier, no significant change in cell number between control and HS-72 treated cells was noted through out the study, indicating cell viability was maintained (Fig. S3). To discriminate the effect of HS-72 on the early stages of the DENV life cycle, an attachment and entry assay in U937+DC-SIGN cells was performed. Cells were treated with HS-72 and cooled to 4°C. To evaluate the effects of HS-72 on viral attachment DENV was added to chilled cells for 1 hour in the presence and absence of HS-72. Separately, cells were transferred from 4°C to 37°C and incubated for an additional hour to allow for viral entry, plus and minus HS-72. Cells were then processed and analyzed by qPCR to quantify viral RNA. These studies showed that there is a significant decrease in viral RNA at the entry stage of the viral life cycle in cells treated with HS-72 (Fig. 3B). Collectively, these data indicate that HS-72 is working to inhibit infection by perturbing the entry stage of the DENV life cycle. Furthermore, this indicates that Hsp70i plays a role in mediating DENV entry into monocytes.
Fig. 3. HS-72 inhibits DENV infection at the entry stage of the viral life cycle.
(A) HS-72 inhibits DENV infection at early stages of the viral life cycle as determine by a time of addition assay. HS-72 (75 nmol/ml) was added to U937+DC-SIGN cells at the indicated time points (hours), with DENV added at time point 0. At 24 hours post infection, cells were analyzed through Cellomics with a DENV E protein antibody coupled with a fluorescent secondary antibody to determine cells that are positive for infection. This shows that HS-72 works to reduce DENV infection when added 1 hour before, or at the 0 or 1 hour time points. (B) An attachment/entry assay shows that HS-72 is working to inhibit DENV infection at the entry stage of the viral life cycle. U937+DC-SIGN cells were treated with HS-72 (75 nmol/ml) for 1 hour, cells were then cooled to 4°C and DENV was added. A 1 hour incubation at 4°C, corresponds to the attachment stage. Cells incubated at 37°C for an additional hour corresponds to the entry stage of the life cycle. Samples were then analyzed by qPCR and the fold change of viral RNA compared to GAPDH, which served as the control, was determined. This shows a significant decrease in viral RNA at the entry stage of the DENV life cycle.
(A – B) Mean ± SEM. *, p<0.05 compared to control.
See also S3 Fig.
3.4. Hsp70i localizes to the cell surface following DENV infection
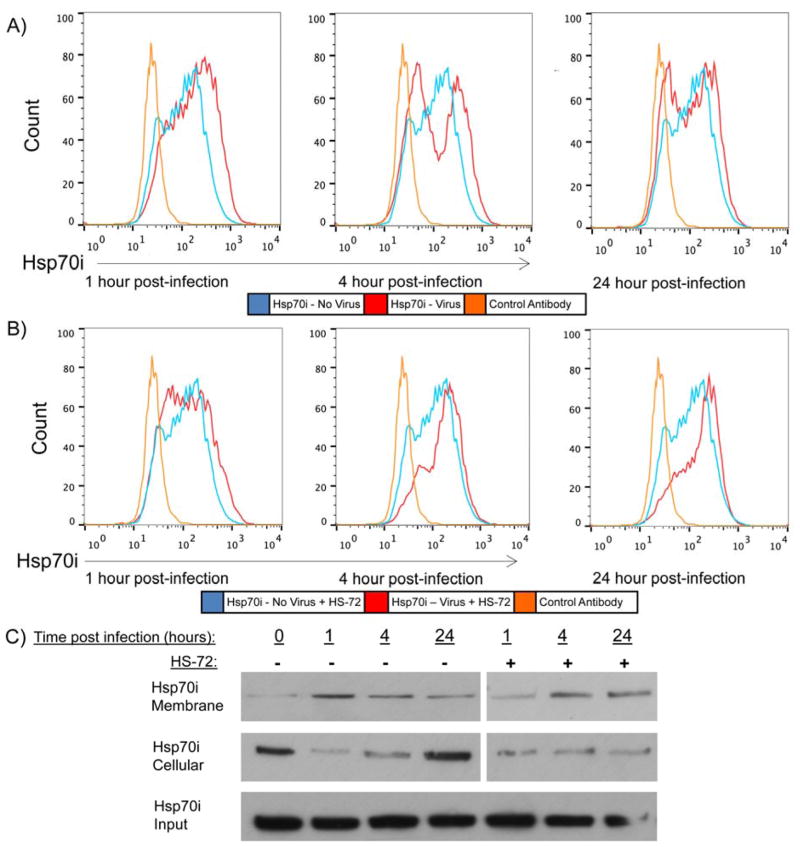
Based on the finding that HS-72 works to disrupt entry of DENV into monocytes, we examined expression of Hsp70i on the cell surface. As discussed earlier, we had noted that DENV infection alone not only induced Hsp70i expression in U937+DC-SIGN cells, but also promoted expression of the protein on the plasma membrane (Figs. 1D and S1D). To further examine this observation, U937+DC-SIGN cells were infected with DENV and analyzed at time points that correlate with pre- and post-entry stages of the DENV life cycle by flow cytometry. Fig. 4A shows there is a significant increase in cell surface Hsp70i 1 hour post-infection, compared to uninfected cells (see also Fig. S4A). However, by 4 and 24 hours post-infection, cell surface Hsp70i has decreased significantly compared to the 1 hour time point (Figs. 4A and S4A). Figure 4A also indicates 2 populations of cells corresponding to Hsp70i expression. It is likely that the cells staining positive for Hsp70i, correspond with the cells that were successfully infected by DENV, while the population that is negative for Hsp70i are cells that were not successfully infected by DENV. Biotin labeling of Hsp70i through its surface cysteine residues on the cell surface further illustrates this change in localization. Isolating the membrane and cytoplasmic fraction of Hsp70i, following biotinylation of the membrane fraction, shows the increase in surface Hsp70i 1 hour post-infection, while there is a decrease in cytoplasmic Hsp70i compared to uninfected cells (Fig. 4C). By 24 hours post-infection, Hsp70i on the cell surface has decreased and there is a commensurate increase in cytoplasmic Hsp70i (Fig. 4C). This data indicates a change in Hsp70i localization following DENV infection to mediate virus entry into U937+DC-SIGN cells. Next, Hsp70i surface expression was examined in DENV infected U937-DC+SIGN cells treated with HS-72, at the same time points as previously described. There is also an observed increase in cell surface Hsp70i expression 1 hour post-infection in HS-72 treated cells compared to uninfected cells (Figs. 4B and S4B). However, by 4 hours and 24 hours post-infection, Hsp70i is continually maintained on the cell surface of HS-72 treated cells (Figs. 4B and S4B). This change in Hsp70i localization is further illustrated through biotinylation of the membrane fraction of Hsp70i in infected cells treated with HS-72, showing Hsp70i maintained on the cell surface by 24 hours post-infection (Fig. 4C). Importantly, HS-72 alone does not change Hsp70i surface expression in uninfected U937-DC+SIGN cells, indicating the change in Hsp70i surface expression is mediated by infection and not by HS-72 treatment alone (Fig. S4C). Furthermore, U937+DC-SIGN cells treated with inactivated DENV, which was heated at 55°C for 30 minutes prior to treatment, show no change in cell surface Hsp70i compared to no virus at the indicated time points, in cells treated with or without HS-72 (Figs. S4D–G). Inactivation of DENV results in no infection of U937+DC-SIGN cells (Fig. S4H). This indicates that the change in Hsp70i localization is mediated by successful infection of U937+DC-SIGN cells. Collectively, this data indicate upon DENV binding and subsequent infection of U937+DC-SIGN cells there is a change in Hsp70i localization to the cell surface. Inhibition of Hsp70i ATPase activity and a conformation change induced by HS-72, maintains Hsp70i on the cell surface in infected cells, but blocks viral entry, suggesting that fully functional Hsp70i is required for entry of DENV into host cells.
Fig. 4. Hsp70i localizes to the cell surface following DENV infection.
(A) Hsp70i surface expression was determined at the indicated time points post-infection, showing a significant increase 1 hour post-infection compared to uninfected cells. While, Hsp70i surface expression has significantly decreased by 4 and 24 hours post-infection. U937+DC-SIGN cells were infected with DENV and at the indicated time points post infection cells were processed for flow cytometry and incubated with a Hsp70i antibody and fluorescent secondary antibody. Fluorescence was measured, which was used to determine Hsp70i surface expression. (B) Hsp70i surface expression was determined in HS-72 (75 nmol/ml) treated cells at the indicated time points post-infection by flow cytometry, showing a significant increase 1 hour, 4 hours, and 24 hours post-infection compared to uninfected cells. This indicates that inhibiting Hsp70i ATPase activity and inducing a conformational change with HS-72 maintains Hsp70i on the cell surface following DENV infection. Cells infected and analyzed as described in (A). (C) Biotinylation of surface Hsp70i shows Hsp70i localizing to the cell surface following DENV infection. While in DENV infected cells treated with HS-72, Hsp70i is maintained on the cell surface until 24 hours post-infection. U937+DC-SIGN cells were infected with DENV and treated with HS-72 (75 nmol/ml) where indicated. At the specified time points, cells were labeled with Sulfo-NHS-SS-Biotin, and incubated at 4°C. Free bio tin was quenched, cells were lysed, and bound to an avidin-sepharose resin. The resin was washed and bound protein was eluted with 2× sample buffer. This resulted in the membrane bound fraction. The cellular fraction was protein that remained unbound after incubation with the avidin-sepharose resin. Input serves as the loading control.
See also S4 Fig.
3.5. Hsp70i interacts with the DENV receptor complex, which is disrupted by HS-72
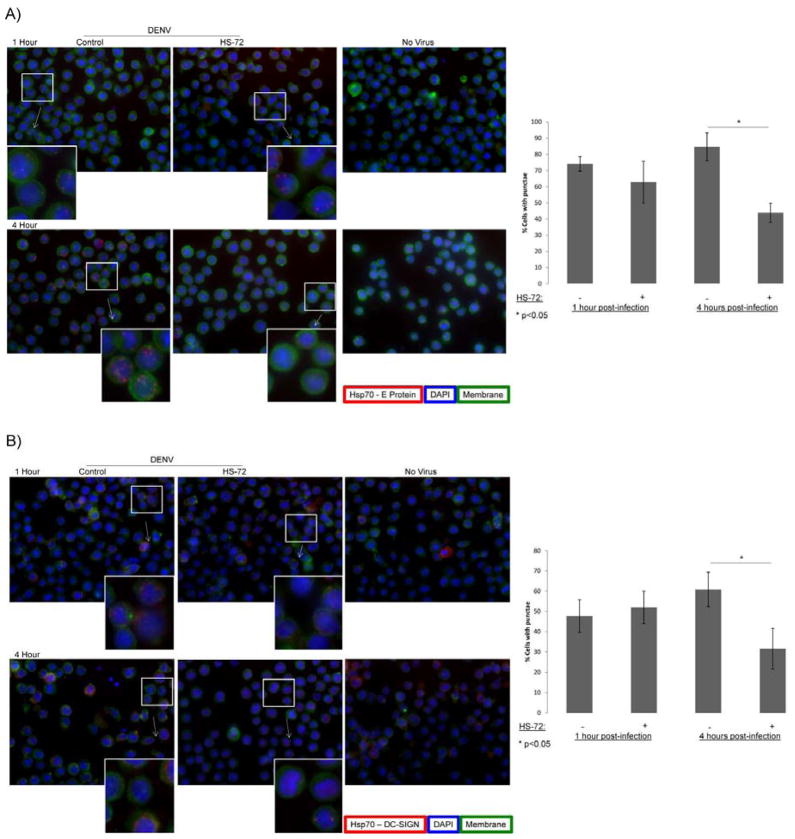
To further elucidate how Hsp70i is mediating DENV entry an interaction between Hsp70i and the DENV receptor complex was investigated through proximity ligation assay (PLA). This assay allows for visualization of in situ interactions in cells by fluorescence microscopy (38). In PLA, primary antibodies were added to cells, followed by addition of PLA probes, which are conjugated with complementary oligonucleotides. Next the probes were hybridized to form a closed circle, which serves as the template for rolling circle amplification (RCA). Addition of fluorescently labeled oligonucleotides, which hybridized the RCA product, yields a fluorescent signal. This signal is visualized by fluorescence microscopy, representing an in situ interaction between the proteins of interest. Fig. 5A shows distinct red punctae were observed, indicating an in situ interaction between Hsp70i and the DENV E protein at 1 and 4 hours post-infection. HS-72 treatment reduced the number of puncta, suggesting a disruption of the interaction at 4 hours post-infection (Figs. 5A). Uninfected cells or cells not treated with primary antibody show very little background fluorescence or red punctae, indicating the specificity of PLA (Figs. 5A and S5). An in situ interaction of Hsp70i with the DC-SIGN receptor was also observed by PLA (Fig. 5B). DC-SIGN has been previously shown to be part of the DENV receptor complex that mediates DENV binding to host cells (39). PLA shows Hsp70i interacting with DC-SIGN at 1 and 4 hours post-infection, while HS-72 also disrupts this interaction 4 hours post-infection (Figs. 5B). An interaction is also observed in uninfected cells, because Hsp70i and DC-SIGN are host proteins (Fig. 5B). Collectively this data shows an in cell interaction of Hsp70i with the DENV E protein and Hsp70i with DC-SIGN, suggesting an interaction of Hsp70i with the DENV receptor complex during DENV binding and entry. HS-72 treatment results in disruption of Hsp70i associating with the DENV receptor complex to perturb entry of DENV into host cells, which ultimately results in a reduction of infected cells.
Fig. 5. Proximity Ligation Assay illustrates an interaction of Hsp70i with the DENV receptor complex, which is disrupted by HS-72, yielding an inhibition in viral entry.
(A) Proximity Ligation Assay (PLA) shows an in situ interaction of Hsp70i with the DENV E protein, which is disrupted by HS-72. U937+DC-SIGN cells were treated with HS-72 (75 nmol/ml) for 1 hour, infected with DENV, and processed for PLA at the indicated time points. Cells were adhered to cover slips, incubated with WGA-488 to stain membranes, blocked and incubated with the indicated primary antibodies. PLA probes were added, ligated, and amplified, cells were then stained with DAPI and mounted for imaging. Fluorescence microscopy shows distinct red punctae, which indicate the in situ interaction of Hsp70i and the DENV E protein at 1 and 4 hours post-infection. The Hsp70i - E protein in situ interaction is disrupted in cells treated with HS-72 at 4 hours post-infection, as observed by a decrease in red punctae. No virus controls shows no punctae, indicating the specificity of the Hsp70i - E protein interaction. Graph on right represents quantification of images from PLA, highlighting the reduction in the Hsp70i - E protein interaction in cells treated with HS-72 at 4 hours post-infection. The percentage of cells with red punctae was quantified by counting cell number as well as the number of cells with red punctae, indicating an interaction between Hsp70i and the E protein, for 5 separate fields of view. (Mean ± SEM. *, p<0.05 compared to control).
(B) PLA shows an in situ interaction of Hsp70i with DC-SIGN, which is disrupted by HS-72. Cells were treated and samples prepared as described in (A).
Fluorescence microscopy shows distinct red punctae, which indicate the in situ interaction of Hsp70i and DC-SIGN at 1 and 4 hours post-infection. The in situ interaction is disrupted in cells treated with HS-72 at 4 hours post-infection, as observed by a decrease in red punctae. No virus controls also show punctae, because Hsp70i and DC-SIGN are host proteins expressed in the cells. Graph on right represents quantification of images from PLA, highlighting the reduction in the Hsp70i - DC-SIGN interaction in cells treated with HS-72 at 4 hours post-infection. The percentage of cells with red punctae was quantified by counting cell number as well as the number of cells with red punctae, indicating an interaction between Hsp70i and DC-SIGN, for 5 separate fields of view. (Mean ± SEM. *, p<0.05 compared to control).
(A–B) Representative images shown with insets showing magnified portion of image designated by boxes. Hsp70i - E protein or Hsp70i - DC-SIGN interaction represented by red punctae, DAPI shown in blue to represent cell nuclei, green staining represents cell membranes.
See also S5 Fig.
4. Discussion
Because of the limited size of most viral genomes, the concept that viruses require components of the host cellular machinery to replicate is not surprising. However, defining which host factors are essential for viral replication using genetic, gene silencing or molecular biological approaches can often be problematic because these techniques often compromise cellular integrity, promote compensatory adaptations or simply trigger programed cell death by activating apoptotic pathways. The outcome of such studies therefore becomes ambiguous, since any antiviral effect could be the result of a dying host cell. Small molecule inhibitors can greatly enable the role of a host protein to be better defined because they can be added at any stage of the life cycle, acutely or chronically, across cell lines and even species while maintaining cellular architecture. Small molecules generally inhibit biological activity rather than removing or mutating the targeted protein, which leads to unwanted compensatory effects. Ultimately, such studies can lead directly to the development of a new antiviral agent. Importantly host targets are not under the direct genetic control of the virus itself and therefore are not prone to early development of resistance under selective drug pressure. We suggest that our studies herein with HS-72, strongly indicate an essential role for Hsp70i in the DENV infection cycle and validate the protein as an antiviral host target. Data from our studies that support this hypothesis can be summarized as follows; 1) proteomic studies show Hsp70i is induced and trafficked to the plasma membrane in response to DENV infection of the human liver cell line Huh7 and in a monocyte line; 2) pharmacological induction of Hsp70i expression with an Hsp90 inhibitor potently promotes DENV infectivity; 3) HS-72, a highly selective allosteric inhibitor of Hsp70i exhibits anti-DENV activity in both a monocyte and liver cell lines; 4) HS-72 blocks DENV entry by inhibiting Hsp70i at the plasma membrane; 5) cell imaging using the proximity ligation assay (PLA) show co-localization between Hsp70i, DENV E protein and DC SIGN in monocytes and these interactions are disrupted by HS-72. Collectively these findings suggest a model in which DENV, perhaps through interactions with DC-SIGN, triggers induced expression and trafficking of Hsp70i to the plasma membrane followed by binding to the virally encoded E protein. Presumably this triggers ATP turnover or a conformational change in Hsp70i that facilitates reinternalization along with the viral particle to promote infection. Inhibition of Hsp70i by HS-72 prevents Hsp70i reinternalization and disrupts Hsp70i interacting with the DENV receptor complex, therefore blocking viral entry. To test this hypothesis further detailed studies between Hsp70i, DENV E protein and DC SIGN receptor are certainly warranted. However, if this mechanism is correct, Hsp70i could be the second example of a druggable host protein that functions as a cell surface receptor for a virus that is necessary for viral entry. The anti-HIV drug Maraviroc functions to target the CCR5 protein and its interactions with CD4, to block HIV invasion of CD4+ T cells (40).
A recent report shows that Hsp70 family members are involved in all stages of the viral lifecycle, with the specific function mediated by Hsp70 co-chaperones (41). In this work Taguwa, et. al. show that Hsp70 regulates DENV entry and targeting the Hsp70 – co-chaperone interaction with an allosteric inhibitor inhibits DENV entry. This new report correlates with our data, in that Hsp70 works to mediate DENV entry, while our data provides more understanding to the role of the inducible Hsp70 family member. However, Taguwa, et. al. report that their allosteric inhibitors work to block DENV at multiple stages of the viral life cycle, while HS-72 works to block on the entry stage of the viral life cycle. This suggests that targeting Hsp70 at distinct allosteric sites and disrupting the Hsp70 – co-chaperone interactions results in varying effects on the DENV life cycle.
There is great interest in Hsp70i as a drug target for the treatment of various cancers (42, 43). As discussed, Hsp70i belongs to a group of closely related heat shock proteins, that share close sequence identity with the protein, in particular the non-inducible housekeeping protein, Hsc70. The Hsp70 proteins are thought to function in cellular protein homeostasis and are required for normal protein folding and stability. These activities often require ATPase activity and typically, like many heat shock proteins, the Hsp70s all possess ATP binding domains. This feature makes such proteins highly druggable and in cancer much attention has been focused in recent years on developing specific inhibitors for the heat shock proteins. In cancer, inhibition of Hsp90 or Hsp70 results in inhibition of cellular proliferation and metastasis (37, 44). This response is associated with down regulation of the expression of various oncogenic client proteins such as HER2, B Raf and AKT (37, 44). Indeed, the list of signal transduction proteins associated with both Hsp90 and Hsp70 is extensive (37). Our data highlight several interesting parallels between oncogenic transformation and viral infection and their connections with the heat shock proteins. Cancer cells are often described as being addicted to heat shock proteins to maintain cellular proliferation (42). Heat shock proteins such as Hsp90 and Hsp70 are not oncogenes themselves, but are induced and sequestered by oncogenic proteins, promoting their proper folding and stability. Inhibiting Hsp90 or Hsp70 ATPase activity blocks this function, generally putting the cell into dormancy. For these reasons heat shock protein inhibitors are highly synergistic with antibodies or drugs that target the oncogene itself (45, 46). Therefore, the sequestration of Hsp70i by DENV to promote viral entry is analogous to oncogene sequestration of the protein and other Hsps. It is likely therefore, that inhibitors like HS-72, would be highly synergistic with any inhibitors targeting viral encoded nonstructural proteins or antibodies targeting viral surface proteins such as E protein or prM.
Our immediate goal is to improve the potency of HS-72 as an anti-DENV agent. In recent work, we demonstrated that HS-72 has excellent pharmacokinetic and distribution properties in mice accumulating in tissues to [mM], with t1/2 elimination times of >24 hours (Howe et al. 2014). The compound is well tolerated in mice and exhibited efficacy against the MMTV model of human breast cancer. Clearly the HS-72 scaffold has excellent drug like characteristics and should be tested in animal models of DENV infection across serotypes, either as a mono therapy or in combination with NS5 inhibitors (47, 48). Given the synergistic effects often observed with heat shock inhibitors with targeted chemotherapeutics in cancer studies, HS-72 may show synergism with NS5 inhibitors that have failed to progress clinically due to dose limiting toxicities (47, 49, 50). If HS-72 proves itself useful as an antiviral agent, one can draw another parallel between cancer biology and antiviral development. The first clinically relevant antiretroviral drug AZT was originally derived from a cancer drug discovery program (51). In the case of AZT, the drug had dose limiting toxicities in cancer that limited its usefulness (51). However, its potency towards the HIV encoded reverse transcriptase greatly expanded its therapeutic window, especially when finally used in combination with protease inhibitors (51).
4.1. Conclusion
Due to the relatively small size of viral genomes, host factors are utilized throughout the viral life cycle. We have identified an essential role for the inducible Heat Shock Protein 70 (Hsp70i) in the Dengue Virus (DENV) life cycle. DENV is of great public health importance due to estimates of up to 400 million infections per year coupled with the geographic distribution of the virus, which is endemic in over 100 countries worldwide. Additionally, there is a pressing need for DENV interventions owing to the lack of approved vaccines or antiviral therapies. We have previously identified a small molecule inhibitor, called HS-72, which is a selective allosteric inhibitor for Hsp70i. Studies presented here with HS-72 have revealed a potentially indispensable role of Hsp70i in DENV pathogenesis and validate Hsp70i as a host target for anti-DENV treatments. Antiviral activity of HS-72 is exerted during the viral entry stage, through disruption of Hsp70i interacting with the DENV receptor complex. The HS-72 scaffold has excellent pharmacological properties and presents a viable starting point for further development of antiviral inhibitors targeting Hsp70i.
Supplementary Material
The stress inducible Hsp70 family member (Hsp70i) is a dengue virus (DENV) host factor.
HS-72, an allosteric small molecule inhibitor selective for Hsp70i reduces DENV infection.
Hsp70i localizes to the cell surface and associates with the DENV receptor complex to mediate entry into host cells.
HS-72 blocks entry of DENV by disrupting the association of Hsp70i with the DENV receptor complex.
This work highlights the potential of the HS-72 scaffold as an antiviral agent for DENV infection.
Acknowledgments
Special thanks to Dr. Mariano Garcia-Blanco for providing DENV antibodies and DENV stocks as well as helpful discussions. The structure and use of the HS-72 scaffold and its resynthesis have been disclosed to Duke University in accordance with Duke University guidelines concerning potential intellectual property. This work was funded by NIH grant R01-AI089526-04 to TAJH and a Duke/Duke-NUS Collaborative grant to TAJH and SV.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Guzman MG, Halstead SB, Artsob H, Buchy P, Farrar J, Gubler DJ, et al. Dengue: a continuing global threat. Nature reviews Microbiology. 2010;8(12 Suppl):S7–16. doi: 10.1038/nrmicro2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kyle JL, Harris E. Global spread and persistence of dengue. Annual review of microbiology. 2008;62:71–92. doi: 10.1146/annurev.micro.62.081307.163005. [DOI] [PubMed] [Google Scholar]
- 3.Wilder-Smith A, Ooi EE, Vasudevan SG, Gubler DJ. Update on dengue: epidemiology, virus evolution, antiviral drugs, and vaccine development. Current infectious disease reports. 2010;12(3):157–64. doi: 10.1007/s11908-010-0102-7. [DOI] [PubMed] [Google Scholar]
- 4.Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, et al. The global distribution and burden of dengue. Nature. 2013;496(7446):504–7. doi: 10.1038/nature12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Back AT, Lundkvist A. Dengue viruses - an overview. Infection ecology & epidemiology. 2013;3 doi: 10.3402/iee.v3i0.19839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coller BA, Clements DE. Dengue vaccines: progress and challenges. Current opinion in immunology. 2011;23(3):391–8. doi: 10.1016/j.coi.2011.03.005. [DOI] [PubMed] [Google Scholar]
- 7.Whitehead SS, Blaney JE, Durbin AP, Murphy BR. Prospects for a dengue virus vaccine. Nature reviews Microbiology. 2007;5(7):518–28. doi: 10.1038/nrmicro1690. [DOI] [PubMed] [Google Scholar]
- 8.Halstead SB, Nimmannitya S, Cohen SN. Observations related to pathogenesis of dengue hemorrhagic fever. IV. Relation of disease severity to antibody response and virus recovered. The Yale journal of biology and medicine. 1970;42(5):311–28. [PMC free article] [PubMed] [Google Scholar]
- 9.Thomas SJ, Endy TP. Critical issues in dengue vaccine development. Current opinion in infectious diseases. 2011;24(5):442–50. doi: 10.1097/QCO.0b013e32834a1b0b. [DOI] [PubMed] [Google Scholar]
- 10.Morrison AC, Zielinski-Gutierrez E, Scott TW, Rosenberg R. Defining challenges and proposing solutions for control of the virus vector Aedes aegypti. PLoS medicine. 2008;5(3):e68. doi: 10.1371/journal.pmed.0050068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ross TM. Dengue virus. Clinics in laboratory medicine. 2010;30(1):149–60. doi: 10.1016/j.cll.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ang F, Wong AP, Ng MM, Chu JJ. Small interference RNA profiling reveals the essential role of human membrane trafficking genes in mediating the infectious entry of dengue virus. Virology journal. 2010;7:24. doi: 10.1186/1743-422X-7-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fernandez-Garcia MD, Mazzon M, Jacobs M, Amara A. Pathogenesis of flavivirus infections: using and abusing the host cell. Cell host & microbe. 2009;5(4):318–28. doi: 10.1016/j.chom.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 14.Sessions OM, Barrows NJ, Souza-Neto JA, Robinson TJ, Hershey CL, Rodgers MA, et al. Discovery of insect and human dengue virus host factors. Nature. 2009;458(7241):1047–50. doi: 10.1038/nature07967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Noble CG, Chen YL, Dong H, Gu F, Lim SP, Schul W, et al. Strategies for development of Dengue virus inhibitors. Antiviral research. 2010;85(3):450–62. doi: 10.1016/j.antiviral.2009.12.011. [DOI] [PubMed] [Google Scholar]
- 16.Chavez-Salinas S, Ceballos-Olvera I, Reyes-Del Valle J, Medina F, Del Angel RM. Heat shock effect upon dengue virus replication into U937 cells. Virus research. 2008;138(1–2):111–8. doi: 10.1016/j.virusres.2008.08.012. [DOI] [PubMed] [Google Scholar]
- 17.Padwad YS, Mishra KP, Jain M, Chanda S, Ganju L. Dengue virus infection activates cellular chaperone Hsp70 in THP-1 cells: downregulation of Hsp70 by siRNA revealed decreased viral replication. Viral immunology. 2010;23(6):557–65. doi: 10.1089/vim.2010.0052. [DOI] [PubMed] [Google Scholar]
- 18.Reyes-Del Valle J, Chavez-Salinas S, Medina F, Del Angel RM. Heat shock protein 90 and heat shock protein 70 are components of dengue virus receptor complex in human cells. Journal of virology. 2005;79(8):4557–67. doi: 10.1128/JVI.79.8.4557-4567.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Daugaard M, Rohde M, Jaattela M. The heat shock protein 70 family: Highly homologous proteins with overlapping and distinct functions. FEBS letters. 2007;581(19):3702–10. doi: 10.1016/j.febslet.2007.05.039. [DOI] [PubMed] [Google Scholar]
- 20.Tavaria M, Gabriele T, Kola I, Anderson RL. A hitchhiker’s guide to the human Hsp70 family. Cell stress & chaperones. 1996;1(1):23–8. doi: 10.1379/1466-1268(1996)001<0023:ahsgtt>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brenner BG, Wainberg MA. Heat shock protein-based therapeutic strategies against human immunodeficiency virus type 1 infection. Infectious diseases in obstetrics and gynecology. 1999;7(1–2):80–90. doi: 10.1155/S1064744999000150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheung RK, Dosch HM. The growth transformation of human B cells involves superinduction of hsp70 and hsp90. Virology. 1993;193(2):700–8. doi: 10.1006/viro.1993.1178. [DOI] [PubMed] [Google Scholar]
- 23.Lefeuvre A, Contamin H, Decelle T, Fournier C, Lang J, Deubel V, et al. Host-cell interaction of attenuated and wild-type strains of yellow fever virus can be differentiated at early stages of hepatocyte infection. Microbes and infection/Institut Pasteur. 2006;8(6):1530–8. doi: 10.1016/j.micinf.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 24.Liao WJ, Fan PS, Fu M, Fan XL, Liu YF. Increased expression of 70 kD heat shock protein in cultured primary human keratinocytes induced by human papillomavirus 16 E6/E7 gene. Chinese medical journal. 2005;118(24):2058–62. [PubMed] [Google Scholar]
- 25.Mayer MP. Recruitment of Hsp70 chaperones: a crucial part of viral survival strategies. Reviews of physiology, biochemistry and pharmacology. 2005;153:1–46. doi: 10.1007/s10254-004-0025-5. [DOI] [PubMed] [Google Scholar]
- 26.Vega-Almeida TO, Salas-Benito M, De Nova-Ocampo MA, Del Angel RM, Salas-Benito JS. Surface proteins of C6/36 cells involved in dengue virus 4 binding and entry. Arch Virol. 2013;158(6):1189–207. doi: 10.1007/s00705-012-1596-0. [DOI] [PubMed] [Google Scholar]
- 27.Howe MK, Bodoor K, Carlson DA, Hughes PF, Alwarawrah Y, Loiselle DR, et al. Identification of an allosteric small-molecule inhibitor selective for the inducible form of heat shock protein 70. Chemistry & biology. 2014;21(12):1648–59. doi: 10.1016/j.chembiol.2014.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nature methods. 2012;9(7):671–5. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gabriel L, Stevens Z, Melikian H. Measuring plasma membrane protein endocytic rates by reversible biotinylation. Journal of visualized experiments : JoVE. 2009;(34) doi: 10.3791/1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hughes PF, Barrott JJ, Carlson DA, Loiselle DR, Speer BL, Bodoor K, et al. A highly selective Hsp90 affinity chromatography resin with a cleavable linker. Bioorg Med Chem. 2012;20(10):3298–305. doi: 10.1016/j.bmc.2012.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barrott JJ, Hughes PF, Osada T, Yang XY, Hartman ZC, Loiselle DR, et al. Optical and radioiodinated tethered Hsp90 inhibitors reveal selective internalization of ectopic Hsp90 in malignant breast tumor cells. Chemistry & biology. 2013;20(9):1187–97. doi: 10.1016/j.chembiol.2013.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carlson DA, Franke AS, Weitzel DH, Speer BL, Hughes PF, Hagerty L, et al. Fluorescence linked enzyme chemoproteomic strategy for discovery of a potent and selective DAPK1 and ZIPK inhibitor. ACS chemical biology. 2013;8(12):2715–23. doi: 10.1021/cb400407c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haystead TA. The purinome, a complex mix of drug and toxicity targets. Current topics in medicinal chemistry. 2006;6(11):1117–27. doi: 10.2174/156802606777812059. [DOI] [PubMed] [Google Scholar]
- 34.Fadden P, Huang KH, Veal JM, Steed PM, Barabasz AF, Foley B, et al. Application of chemoproteomics to drug discovery: identification of a clinical candidate targeting hsp90. Chemistry & biology. 2010;17(7):686–94. doi: 10.1016/j.chembiol.2010.04.015. [DOI] [PubMed] [Google Scholar]
- 35.Guo F, Rocha K, Bali P, Pranpat M, Fiskus W, Boyapalle S, et al. Abrogation of heat shock protein 70 induction as a strategy to increase antileukemia activity of heat shock protein 90 inhibitor 17-allylamino-demethoxy geldanamycin. Cancer research. 2005;65(22):10536–44. doi: 10.1158/0008-5472.CAN-05-1799. [DOI] [PubMed] [Google Scholar]
- 36.Powers MV, Workman P. Inhibitors of the heat shock response: biology and pharmacology. FEBS letters. 2007;581(19):3758–69. doi: 10.1016/j.febslet.2007.05.040. [DOI] [PubMed] [Google Scholar]
- 37.Powers MV, Clarke PA, Workman P. Death by chaperone: HSP90, HSP70 or both? Cell cycle. 2009;8(4):518–26. doi: 10.4161/cc.8.4.7583. [DOI] [PubMed] [Google Scholar]
- 38.Koos B, Andersson L, Clausson CM, Grannas K, Klaesson A, Cane G, et al. Analysis of protein interactions in situ by proximity ligation assays. Curr Top Microbiol Immunol. 2014;377:111–26. doi: 10.1007/82_2013_334. [DOI] [PubMed] [Google Scholar]
- 39.Tassaneetrithep B, Burgess TH, Granelli-Piperno A, Trumpfheller C, Finke J, Sun W, et al. DC-SIGN (CD209) mediates dengue virus infection of human dendritic cells. The Journal of experimental medicine. 2003;197(7):823–9. doi: 10.1084/jem.20021840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dorr P, Westby M, Dobbs S, Griffin P, Irvine B, Macartney M, et al. Maraviroc (UK-427,857), a potent, orally bioavailable, and selective small-molecule inhibitor of chemokine receptor CCR5 with broad-spectrum anti-human immunodeficiency virus type 1 activity. Antimicrobial agents and chemotherapy. 2005;49(11):4721–32. doi: 10.1128/AAC.49.11.4721-4732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Taguwa S, Maringer K, Li X, Bernal-Rubio D, Rauch JN, Gestwicki JE, et al. Defining Hsp70 Subnetworks in Dengue Virus Replication Reveals Key Vulnerability in Flavivirus Infection. Cell. 2015;163(5):1108–23. doi: 10.1016/j.cell.2015.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jego G, Hazoume A, Seigneuric R, Garrido C. Targeting heat shock proteins in cancer. Cancer letters. 2013;332(2):275–85. doi: 10.1016/j.canlet.2010.10.014. [DOI] [PubMed] [Google Scholar]
- 43.Powers MV, Jones K, Barillari C, Westwood I, van Montfort RL, Workman P. Targeting HSP70: the second potentially druggable heat shock protein and molecular chaperone? Cell cycle. 2010;9(8):1542–50. doi: 10.4161/cc.9.8.11204. [DOI] [PubMed] [Google Scholar]
- 44.Powers MV, Clarke PA, Workman P. Dual targeting of HSC70 and HSP72 inhibits HSP90 function and induces tumor-specific apoptosis. Cancer cell. 2008;14(3):250–62. doi: 10.1016/j.ccr.2008.08.002. [DOI] [PubMed] [Google Scholar]
- 45.Leow CC, Chesebrough J, Coffman KT, Fazenbaker CA, Gooya J, Weng D, et al. Antitumor efficacy of IPI-504, a selective heat shock protein 90 inhibitor against human epidermal growth factor receptor 2-positive human xenograft models as a single agent and in combination with trastuzumab or lapatinib. Mol Cancer Ther. 2009;8(8):2131–41. doi: 10.1158/1535-7163.MCT-08-1038. [DOI] [PubMed] [Google Scholar]
- 46.Wainberg ZA, Anghel A, Rogers AM, Desai AJ, Kalous O, Conklin D, et al. Inhibition of HSP90 with AUY922 induces synergy in HER2-amplified trastuzumab-resistant breast and gastric cancer. Mol Cancer Ther. 2013;12(4):509–19. doi: 10.1158/1535-7163.MCT-12-0507. [DOI] [PubMed] [Google Scholar]
- 47.Lim SP, Wang QY, Noble CG, Chen YL, Dong H, Zou B, et al. Ten years of dengue drug discovery: progress and prospects. Antiviral research. 2013;100(2):500–19. doi: 10.1016/j.antiviral.2013.09.013. [DOI] [PubMed] [Google Scholar]
- 48.Schul W, Liu W, Xu HY, Flamand M, Vasudevan SG. A dengue fever viremia model in mice shows reduction in viral replication and suppression of the inflammatory response after treatment with antiviral drugs. The Journal of infectious diseases. 2007;195(5):665–74. doi: 10.1086/511310. [DOI] [PubMed] [Google Scholar]
- 49.Lai CH, Park KS, Lee DH, Alberobello AT, Raffeld M, Pierobon M, et al. HSP-90 inhibitor ganetespib is synergistic with doxorubicin in small cell lung cancer. Oncogene. 2014;33(40):4867–76. doi: 10.1038/onc.2013.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McConnell JR, McAlpine SR. Heat shock proteins 27, 40, and 70 as combinational and dual therapeutic cancer targets. Bioorg Med Chem Lett. 2013;23(7):1923–8. doi: 10.1016/j.bmcl.2013.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Broder S. The development of antiretroviral therapy and its impact on the HIV-1/AIDS pandemic. Antiviral research. 2010;85(1):1–18. doi: 10.1016/j.antiviral.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.