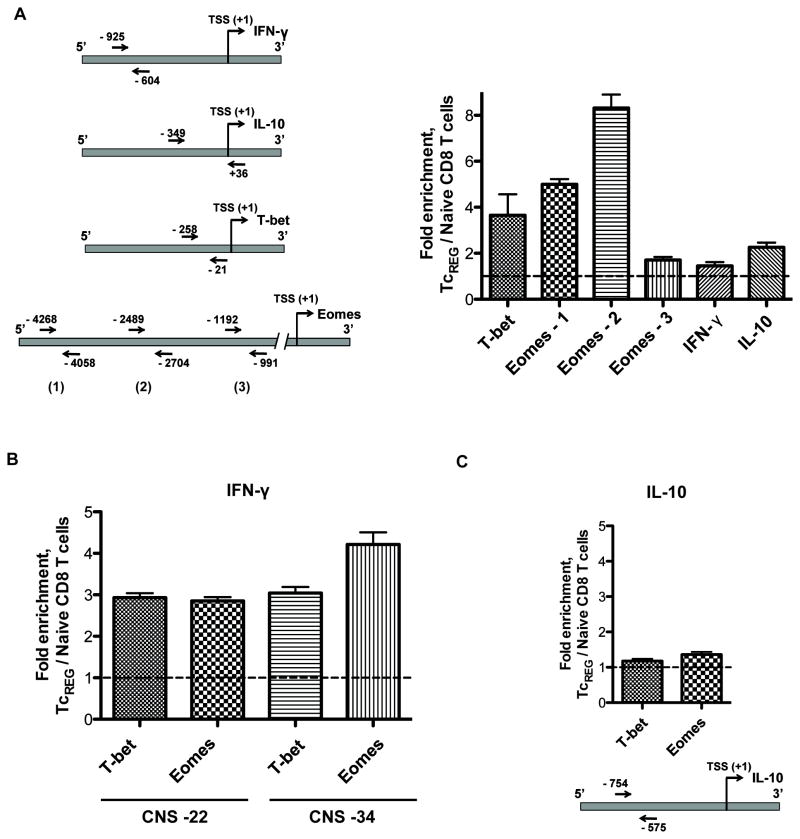
Figure 4. Gene regulatory network.
(A) Measurements of ccupancy of the p65 subunit of NF-κB on T-bet, Eomes, and IL-10 promoters in TcREG cells. Left panel: A diagram showing location of primers used for qPCR to detect enrichment of p65-binding sites. Right panel: Enrichment of p65-binding DNA sequences in the promoters of indicated genes of TcREG cells relative to naïve CD8 T cells. TcREG chromatin for use in ChIP was prepared after 24 h of incubation with osteoclasts. (B) T-bet and Eomes bind to distal conserved sequences CNS-22 and CNS-34 of IFN-γ gene in TcREG. Chromatin from naïve CD8 T cells (negative control) and from 24 h TcREG was immunoprecipitated with anti-T-bet or anti-Eomes antibody. Enrichment of T-bet and Eomes binding sites at −22k and −34K of IFN-γ gene was assessed by qPCR. (C) T-bet and Eomes do not bind IL-10 promoter. Chromatin prepared and immunoprecipitated as described in (B) was subjected to qPCR with primers, which flank putative T-bet/Eomes binding sites in IL-10 promoter. These results show no significant enrichment for these sites in TcREG cells relative to naïve CD8 T cells.