Abstract
The Hsp90 chaperone is a central node of protein homeostasis activating a large number of diverse client proteins. Hsp90 functions as a molecular clamp that closes and opens in response to the binding and hydrolysis of ATP. Crystallographic studies define distinct conformational states of the mechanistic core implying structural changes that have not yet been observed in solution. Here, we engineered one-nanometer fluorescence probes based on photo-induced electron transfer into yeast Hsp90 to observe these motions. We found that the ATPase activity of the chaperone was reflected in the kinetics of specific structural rearrangements at remote positions that acted cooperatively. Nanosecond single-molecule fluorescence fluctuation analysis uncovered that critical structural elements that undergo rearrangement are mobile on a sub-millisecond time scale. We identified a two-step mechanism for lid closure over the nucleotide-binding pocket. The activating co-chaperone Aha1 mobilizes the lid of apo Hsp90, suggesting an early role in the catalytic cycle.
The 90-kDa heat shock protein (Hsp90) is a highly abundant and evolutionary conserved molecular chaperone that acts at the late stages of folding where it stabilizes and activates a plethora of structurally and functionally diverse client proteins. Many of them are essential for signal-transduction such as steroid hormone receptors, kinases, transcription factors as well as viral proteins1–3. Hsp90 is implicated in malignant disease where its chaperone activity is responsible for the stability of several key oncoproteins and is thus a current target of anti-cancer drug development4–6.
Hsp90 is a homo-dimer where each monomer consists of three distinct domains. The N-terminal domain (NTD) contains the ATP-binding pocket7, which is also the binding site for the many Hsp90 inhibitors currently in clinical development8–10. The NTD is connected to the middle domain (MD), which is implicated in client protein binding11,12, via a long and flexible charged linker. Constitutive dimerization is provided by the C-terminal domain (CTD)13,14.
Hsp90 undergoes a conformational cycle coupled to a very slow inherent ATPase activity, with time constants of the order of minutes15, which drives transient association of the two NTDs in the dimer16. This ATPase-coupled mechanism is critical to the biological function of Hsp90, and mutational disruption or pharmacological inhibition abolishes its molecular chaperone activity in vivo15,17,18.
Comparison of Hsp90 crystal structures has defined the conformational differences between the relaxed apo or ADP-bound state of the chaperone, in which the NTDs are not constrained to interact with each other, and the closed ‘tense’ state engendered by binding of ATP7,14,19,20. Inter-subunit dimerization of the NTDs and their close juxtaposition to the MDs in the ATP-bound state is controlled by a set of localized conformational switches, driven by the behavior of a critical structural element in the NTD termed the ‘lid’. The lid closes over the nucleotide-binding pocket, trapping the bound ATP while simultaneously exposing a hydrophobic surface that facilitates N-terminal dimerization and exchange of the N-terminal β-strands between the interacting NTDs14,21. Full activation of the ATPase activity requires the additional docking of the MDs onto the NTDs and remodeling of a catalytic loop on the MD to allow a critical arginine (Arg 380 in yeast Hsp90) to project into the top of the nucleotide pocket and interact with the γ-phosphate of ATP11,21. The dramatic effects of mutations in the lid segment on ATPase activity16 suggests that it is restructuring and the connected conformational changes in other parts of the protein, rather than ATP hydrolysis itself, that limits the rate constant of the chaperone catalytic cycle20,21. This model is supported by a number of more recent biophysical studies22–24.
Hsp90 is regulated by a number of collaborating proteins21, so-called co-chaperones, including Hop/Sti1, Cdc37, and p23/Sba1, which all inhibit the ATPase cycle, and Aha1, which in contrast activates Hsp9025. Structural studies of co-chaperone complexes with Hsp9014,26,27 show that these proteins exert much of their regulatory effect by interacting with those segments of the NTD and MD that undergo local structural changes as part of the ATPase mechanism20–24.
To date, our understanding of the structural changes that accompany the ATPase mechanism of Hsp90 has been mainly deduced from crystal structures and biochemical studies. Direct observation of local conformational switching in solution has been hampered by the lack of suitable spectroscopic probes.
Quenching of extrinsic fluorophores by the amino acid tryptophan (Trp) through photo-induced electron transfer (PET) can measure local conformational changes at a scale of ~1 nm28,29. The combination of PET with nanosecond single-molecule fluorescence fluctuation analysis can probe rapid protein folding dynamics30–32. The 1-nm resolution of PET complements the popular fluorescence resonance energy transfer (FRET) approach, which is active on a 2-10 nm scale, in the exploration of protein conformation33.
Here, we engineered PET-based reporter systems into yeast Hsp90 to site-specifically probe kinetics of local conformational changes. We found that the rate constant of ATP hydrolysis was reflected in the rate constants of lid closure, β-strand swap, and intra-subunit association of NTD and MD. These conformational changes, which form the catalytically active unit of Hsp90, appeared to cooperate. We identified a two-step mechanism for lid-closure and a previously unknown mode of action of the co-chaperone Aha1.
Results
Design of fluorescence probes for conformational changes
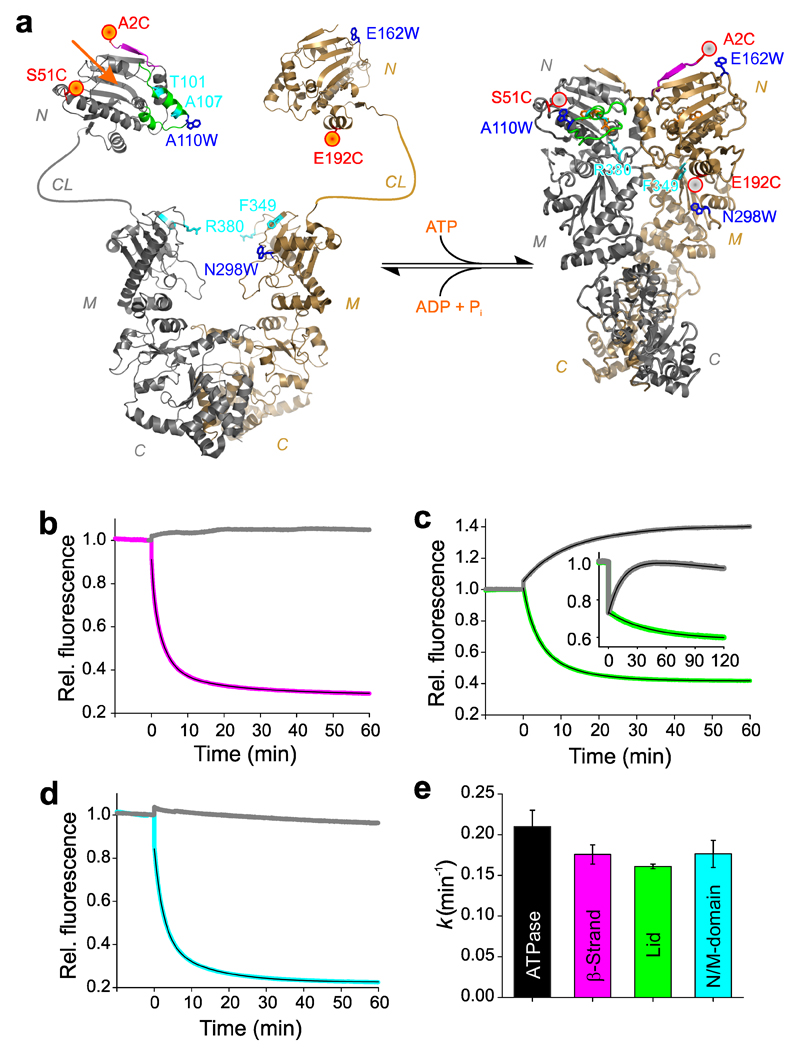
We developed reporter systems based on fluorescence quenching by PET to probe dynamics of local structural changes of Hsp90. PET requires van der Waals contact between an organic fluorophore and the indole side chain of Trp, which occurs at a distance of ≤1 nm34. Based on crystal structures of the closed yeast Hsp90 conformation (pdb id 2CG9) and the isolated NTD (pdb id 1AM1), which was used as a model for the open-clamp NTD conformation, we engineered PET reporters at surface-exposed positions (Figure 1a). We placed Trp and the extrinsic oxazine fluorophore AttoOxa11 (Oxa) such that a change of conformation resulted in formation or disruption of the Oxa-Trp interaction, so that fluorescence was either off or on, respectively. To this end, Trp and cysteine (Cys, C) residues were introduced by site-directed mutagenesis. Cys provided the attachment site for the thiol-reactive derivative of Oxa. Yeast Hsp90 has no inherent Cys that would interfere with site-specific modification. We probed closure of the lid using two PET reporter systems, namely S51C-A110W and A110C-S51W, and intra-subunit association of N- and M-domains using the reporter E192C-N298W. Cross-subunit swap of the N-terminal β-strands (β-strand swap) was monitored by placing Oxa on the N-terminus of one subunit (A2C) and Trp in a waiting-position on the other (E162W).
Figure 1. Observation of conformational motions in Hsp90 by PET fluorescence quenching.
(a) PET reporter design. Left: Structural model of apo Hsp90 based on crystallographic data of the NTD (pdb id 1AM1) and the MC-domain (pdb id 2CGE). NTD (N), charged linker (CL), M-domain (M), and C-domain (C) are indicated. The nucleotide-binding pocket is indicated by an orange arrow. Right: Crystal structure of full-length Hsp90 in closed-clamp conformation with bound AMP-PNP (pdb id 2CG9). N-terminal β-strand and lid are colored magenta and green, respectively. Engineered Oxa and Trp are shown as red spheres and blue sticks, respectively. Amino acid side chains that were mutated to alter function are highlighted in cyan. (b) Fluorescence intensity time traces of reporter A2C-E162W for β-strand swap (magenta) and the corresponding control A2C (gray). AMP-PNP was added at t = 0 min. The black line is a three-exponential fit to the data. (c) Fluorescence intensity time traces of reporter S51C-A110W for lid closure (green) fitted using a bi-exponential function, and of variant S51W-A110C (inset, green). Controls S51C and A110C are shown in gray. (d) Fluorescence intensity time traces of reporter E192C-N298W for N/M-association (cyan) fitted using a bi-exponential function. Control E192C is shown in gray. (e) ATPase activity of wild-type Hsp90 and mean rate constants of β-strand swap, lid closure, and N/M-domain association obtained from PET fluorescence experiments. Data represent mean values ± s.d. of three measurements.
Kinetics of conformational switching of Hsp90
For measurement of intra-subunit conformational changes, i.e. lid-closure and N/M-domain association, we formed hetero-dimers consisting of one wild-type and one PET reporter-containing subunit, thus avoiding complications in data interpretation that would arise from two fluorescence probes located on the same Hsp90 dimer. For measurement of β-strand swap, we formed hetero-dimers consisting of one subunit that contained fluorescently modified A2C and one subunit that contained the E162W mutation. We triggered closure of the molecular clamp by rapidly adding excess of the non-hydrolysable ATP-analogue adenosine 5′-[β,γ-imido]triphosphate (AMP-PNP) to reporter-containing Hsp90 samples and measured the time-dependence of fluorescence intensities in solution. Nucleotide binds to both subunits of Hsp90, which is the physiologically active state35.
All three reporter-systems showed a strong decrease of fluorescence intensity after binding of AMP-PNP (Figure 1b-d). To confirm that the signal changes arose specifically from PET quenching we conducted control measurements with samples that lacked the engineered Trp. The controls showed no fluorescence decrease except for the lid-closure reporter S51C where we observed an increase of fluorescence intensity upon addition of AMP-PNP (Figure 1c). Increase of fluorescence emission may be explained by a change of polarity of the micro-environment of the environmentally sensitive label at position 51 upon structural change. In the second reporter for lid-closure we swapped positions of Trp and Oxa: the label was placed on the rearranging lid segment (A110C) and Trp in waiting position on the opposite site of the nucleotide-binding pocket (S51W). Upon addition of AMP-PNP we observed a rapid drop of fluorescence emission that was faster than the dead time of manual mixing. This burst phase was followed by a slowly decaying signal that resulted from full closure of the lid over the ATP-binding pocket (Figure 1c, inset). The burst phase did not originate from PET quenching since we also observed it in the control sample without engineered Trp, which suggests a rapid change of polarity in the microenvironment of Oxa. Lid closure thus emerged as a two-step process in which binding of nucleotide triggered rapid remodeling of the lid to an intermediate conformation, followed by its slow folding over the binding pocket. Interestingly, some of the many crystal structures of Hsp90 in complex with drugs show partially ordered lid segments, which may be reflective of this intermediate state36. To investigate whether ADP would also induce conformational change in apo Hsp90 we replaced AMP-PNP with ADP in rapid-mixing experiments. ADP did not induce any detectable kinetics in all three reporter systems showing that only the triphosphate derivative was able to trigger switches. However, the burst phase was still present (Supplementary Results, Supplementary Figure 1). This suggested that the nucleotide phosphate group is not required for rapid remodeling of the lid. To gain further insights we resolved the burst phase of the lid using stopped-flow spectroscopy (Supplementary Figure 2). We found that initial remodeling was rate-limited by diffusion-controlled binding of nucleotide. The bi-molecular rate constant, reported by conformational change of the lid, was 1.3±0.2 × 105 M-1 s-1, a value that was in excellent agreement with the one previously obtained from resonance energy transfer experiments using fluorescently modified ATP37.
The slowly decaying fluorescence signals observed for all three conformational switches required a sum of two or three single-exponential functions to describe the data accurately (Supplementary Table 1), which raised the question as to the origin of the observed heterogeneity in kinetics. Structural studies show that the apo-state of Hsp90 is a heterogeneous ensemble of open-clamp conformers38,39. Different conformational ground states each associated with a different free energy will give rise to different rate constants of conformational change along parallel pathways to the closed clamp conformation. If the change of brightness of the label upon transition of Hsp90 from an open-clamp to the closed-clamp conformation is similar for the different ground states, the relative amplitudes of kinetic phases reflect the relative populations of ground states from which the transitions originate. We found that the mean rate constant of each motion, calculated as the inverse of the sum of fitted time constants in a multi-exponential decay weighted by the respective amplitudes, was similar and in good agreement with the kcat for ATP-hydrolysis (~0.2 min-1; Figure 1e). The mean rate constants of conformational switching thus accounted for the ATPase activity of the entire ensemble of Hsp90 molecules.
Cooperation of local motions in the chaperone
To investigate cooperation of and possible allosteric communication between conformational switches, we introduced single point mutations to alter functionality and speed of conformational changes. We looked for allosteric effects by introducing a mutation at one site and measuring its effect on remote sites.
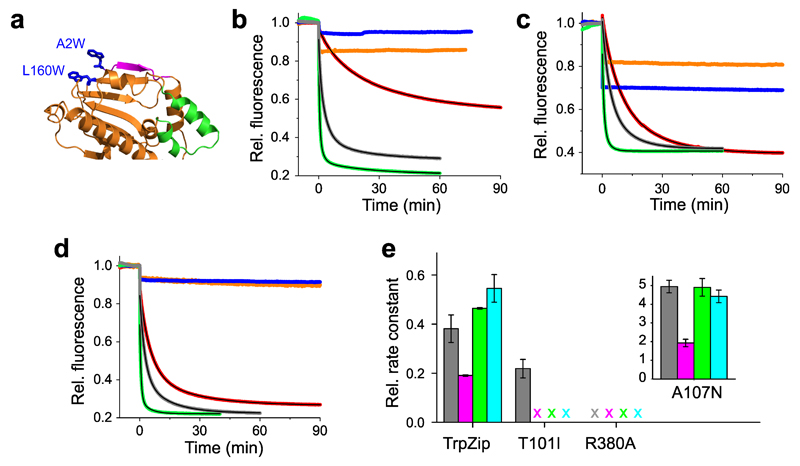
Energetics of β-strand swap was modulated through stabilization of the β-strand on its own domain using a tryptophan-zipper (TrpZip) motif. TrpZip is a designed cross-strand interaction of two Trp side chains on neighboring β-strands, which substantially stabilizes the β-hairpin fold40. TrpZips are widely applied in fundamental studies on mechanisms of β-hairpin folding41. We engineered a TrpZip on the N-terminal β-strand of Hsp90 by mutating Ala2 and Leu160 to Trp (A2W-L160W) (Figure 2a).
Figure 2. Modulation of conformational motions by mutagenesis.
(a) Crystal structure of the NTD (pdb id 1AM1) showing the engineered TrpZip motif introduced through double mutation A2W-L160W (blue sticks). N-terminal β-strand and lid segment are highlighted magenta and green. (b) Time-dependent fluorescence intensities of reporter A2C-E162W (β-strand swap) and mutants thereof. Fluorescence time traces of wild-type (gray), TrpZip (red), mutants T101I (orange), R380A (blue), and A107N (green) are shown. Black lines are exponential fits to the data. AMP-PNP was added at t = 0 min. (c) Time-dependent fluorescence intensities of reporter S51C-A110W (lid) and mutants thereof. The color code of panel (b) applies. (d) Time-dependent fluorescence intensities of reporter E192C-N298W (N/M-domain association) and mutants thereof. The color code of panel (b) applies. (e) Effects of mutation on the rate constant of ATP hydrolysis by Hsp90 (gray) and on the mean rate constants of β-strand swap (magenta), lid closure (green), and N/M-domain association (cyan). ATPase activities of de-activating mutants (TrpZip, T101I, and R380A) were measured at 37° C and plotted as relative rate constants. Data represent mean values ± s.d. of three measurements. X = no kinetics detectable.
To modulate the process of lid-closure we used previously characterized mutations A107N and T101I in the lid segment (Figure 1a). A107N increases ATPase activity about 5-fold through suggested stabilization of the closed-lid conformation14,16, while T101I has the opposite effect, substantially reducing ATPase activity16.
The crystal structure of the closed, N-terminally dimerized Hsp90 shows that Arg380 in the catalytic loop of the M-domain interacts with the γ-phosphate of ATP bound to the NTD and stabilizes an N/M-associated conformation14,42 (Figure 1a). Arg380 provides a connector between the N- and the M-domain and is thought to act as an ATP sensor. Mutation R380A causes severe decrease of ATPase activity in vitro and loss of viability in vivo11. To impair N/M-domain association, we applied mutation R380A.
We introduced each of the modifications described above in Hsp90 constructs containing reporter systems for local motions and measured their effects on AMP-PNP binding-induced kinetics. Fluorescence intensity time traces, data analysis, and ATPase activities are shown in Figure 2. To measure the effect of the TrpZip, we introduced mutation E162C on one subunit and A2W-L160W on the other, and formed hetero-dimers. For the T101I and R380A mutations, which were expected to abolish motion, we applied reporter S51W-A110C that is sensitive to the two-step process of lid closure, as indicated by the presence of a burst phase. Due to the inherently low ATPase activity of Hsp90 at 25 ºC, we conducted ATPase assays at 37 ºC for all deactivating mutants thus increasing assay accuracy. Measured rate constants of ATP hydrolysis and conformational change are provided as Supplementary Tables 1 and 2. Figure 2e shows the relative change of mean rate constant of each motion compared with the relative change of ATPase activity for the corresponding mutation. The TrpZip motif slowed cross-subunit swap of β-strands by ~6-fold. This modification also altered kinetics of N/M-domain association and lid-closure, although to a smaller extent, indicating weak coupling of motions. Mutation A107N, by contrast, accelerated closure of the lid and N/M-domain association by ~5-fold (Figure 2e, inset), in agreement with the predicted stabilizing effect of this mutation14,16. The effect of A107N on β-strand swap was moderate in comparison, in agreement with the weak coupling of β-strand swap with other motions found for the TrpZip construct. Mutations T101I and R380A abolished lid closure, β-strand swap, and N/M-domain association altogether (Figure 2b-d). However, the lid segment still remodeled rapidly as observed by the presence of burst phases (Figure 2c). Burst phases of small amplitude observed for strand swap and N/M-domain association in mutants T101I and R380A may originate from minor sub-populations of rapidly rearranging apo-Hsp90 conformers that are picked up by the environmentally sensitive label. But the amplitudes of these signals were too small to assign a conformational change with confidence or to investigate them further. Mutant R380A was not capable of hydrolyzing ATP, but mutant T101I still showed some residual ATPase activity (Figure 2e). The seemingly conflicting observation of stalled conformational change through mutation T101I but residual ATPase activity can be explained by the fact that Hsp90 molecules that fail to form an N-terminally dimerized state retain small but measurable activities16,43,44.
A more detailed comparison of rate constants and amplitudes of all mutants is provided as Supplementary Figure 3. Analysis showed that the pattern of kinetics was conserved across positions and mutations. An exception was β-strand swap that had an additional exponential phase on a fast time scale, which vanished in the decelerating TrpZip construct but reappeared in the accelerating mutant A107N.
Influence of Aha1 on local motions
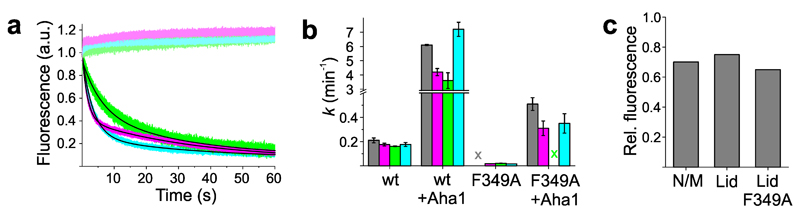
We investigated the influence of the co-chaperone Aha1 on conformational motions in Hsp90. Reporter-containing Hsp90 samples were pre-incubated with Aha1 and motions were triggered by binding of AMP-PNP. Aha1 substantially accelerated conformational changes requiring the use of stopped-flow spectroscopy to measure kinetics on fast time scales. Fluorescence transients showed multi-exponential decays, similar to those observed without Aha1, but on a faster time scale (Figure 3a, Supplementary Table 3). Control measurements of constructs lacking the engineered Trp showed no decays, confirming that the signals of reporter-containing constructs arose specifically from PET fluorescence quenching. The mean rate constant of N/M-domain association was accelerated by ~40 fold, in good agreement with the enhanced ATPase activity, while the mean rate constant of lid-closure and β-strand swap was accelerated by ~20-fold (Figure 3b). Stronger acceleration of N/M-domain association compared with other motions may be explained by pre-association of N- and M-domains induced by Aha123,45. Equilibrium fluorescence intensities of the N/M-domain association reporter showed a significant decrease after binding of Aha1 (Figure 3c), indicating that this preorganization of the N/M-associated state was indeed promoted by Aha1.
Figure 3. Influence of Aha1 on kinetics of local motions.
(a) AMP-PNP-triggered fluorescence intensity time traces of β-strand swap (A2C+E162W, magenta), lid closure (S51C-A110W, green), and N/M-domain association (E192C-N298W, cyan). Samples were incubated with Aha1 before measurement and time traces were recorded using stopped-flow spectroscopy. Data in shaded color are control samples that lacked the engineered Trp. Fluorescence transients were fitted using a bi-exponential model including a linear baseline drift of minor amplitude (black line). (b) Rate constants of ATP hydrolysis (gray) by wild-type Hsp90 (wt) and mutant F349A together with the corresponding mean rate constants of β-strand swap (magenta), lid closure (green), and N/M-domain association (cyan) measured in absence and presence of Aha1 (X = no kinetics detectable). Data represent mean values ± s.d. of three measurements. (c) Equilibrium fluorescence intensities measured from reporters of N/M-domain association and of the lid on wild-type Hsp90 and mutant F349A after incubation with Aha1.
Phenylalanine at position 349 (F349) forms the center of a highly conserved, surface-exposed hydrophobic patch on the M-domain (Figure 1a). This patch is thought to be critical for N/M-association and for correct positioning of the catalytic loop on the M-domain11,45,46. An F349A mutation deactivates Hsp90 and causes dramatic loss of ATPase activity, but activity can be recovered with Aha111,45. We found that Aha1 was indeed capable of stimulating the ATPase activity of F349A to the wild-type level, in agreement with previous observations45 (Figure 3b). Kinetics of N/M-association and β-strand swap of this mutant were similar (Figure 3b, Supplementary Table 3). However, lid reporter S51C-A110W showed fluorescence quenching upon addition of Aha1, both for wild-type and mutant F349A (Figure 3c), suggesting that binding of Aha1 to apo-Hsp90 influenced the lid. As a consequence, the amplitude of AMP-PNP-triggered decay was too small to be fitted accurately.
Early events in the N-terminal domain of Hsp90
Early events occur in the NTD where binding of ATP initiates the chaperone cycle of Hsp90. Remodeling of the lid exposes the dimerization interface that facilitates productive cross-subunit interaction of NTDs and swap of the terminal β-strands. To gain deeper insight into dynamics of lid and β-strand we combined PET with nanosecond single-molecule fluorescence fluctuation analysis, which probes sub-millisecond conformational fluctuations in proteins (PET-FCS)32. In contrast to non-equilibrium rapid-mixing techniques, PET-FCS measures thermally activated equilibrium motions. Fluorescence correlation spectroscopy (FCS) analyzes fluctuations of individual molecules passing through the detection volume of a confocal microscope setup by Brownian motion. Fluorescence time traces are recorded and processed to calculate the second order autocorrelation function (ACF). Besides kinetics of molecular diffusion, protein dynamics are detected from engineered PET reporters that transform conformational fluctuations into fluorescence fluctuations30,32.
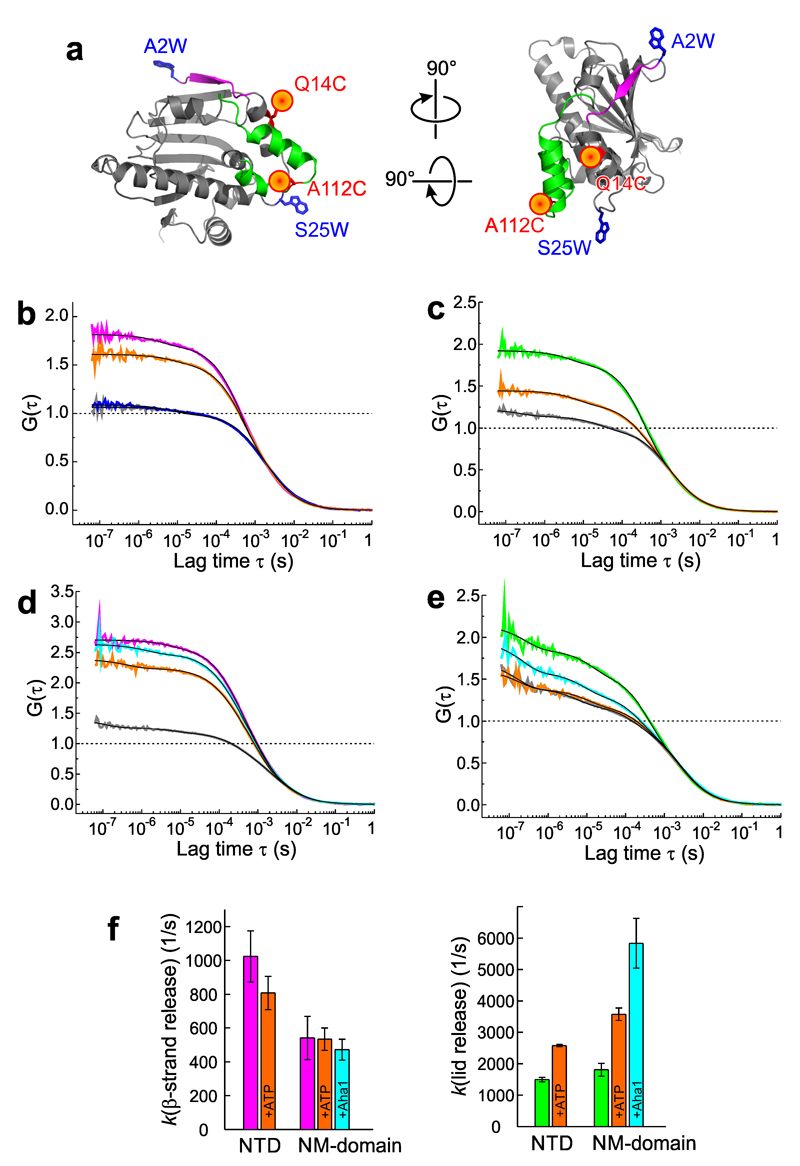
We engineered PET-FCS reporters for dynamics of the N-terminal β-strand and the lid (Figure 4a). Reporter Q14C-A2W was designed to probe β-strand motion through engineering Oxa and Trp on positions that result in fluorescence quenching once the strand was released from the domain. In the folded strand, Oxa and Trp are in 3-nm distance separation such that PET fluorescence quenching cannot occur. Once the β-strand detaches, it forms a mobile coil that facilitates transient interaction of fluorophore and Trp. Reporter A112C-S25W was designed to probe the lid. In A112C-S25W, Oxa and Trp are in PET quenching-distance in the open-lid conformation (Figure 4a). Transient release of a mobile lid from the fully open position would lead to fluorescence fluctuations detected by FCS.
Figure 4. Equilibrium dynamics of lid and β-strand probed by PET-FCS.
(a) Reporter design. N-terminal β-strand and lid of the NTD (pdb id 1AM1) are highlighted magenta and green. Engineered pairs of Oxa (red sphere) and Trp (blue sticks) probing β-strand (Q14C-A2W) and lid (A112C-S25W) are indicated. (b) and (c), ACFs (G(τ)) recorded from Q14C-A2W (magenta) and A112C-S25W (green), respectively, on the isolated NTD. Data recorded after binding of ATP are shown in orange. Control samples lacking the engineered Trp are shown in gray. Black lines are fits to the data using a model for molecular diffusion containing two single-exponential relaxations. The ACF of the TrpZip construct (Q14C-A2W-L60W) is shown in blue in panel (b), and was described by a molecular diffusion model without additional relaxations. All ACFs were normalized to the average number of molecules in the detection focus for clarity. Broken lines indicate the amplitudes of the diffusion decays. (d) and (e) ACFs of same reporters engineered on the NM-domain. Same color code as in panels (b) and (c) applies. Black lines are fits to the data using a model for molecular diffusion containing three single-exponential relaxations. ACFs recorded in the presence of Aha1 are shown in cyan. (f) Rate constants of β-strand release (magenta) and lid release (green) in NTD and NM-domain. Effects of binding of ATP and Aha1 are shown. Data represent mean values ± s.d. of three measurements.
We first investigated the NTD in isolation to study local dynamics without interference from possible intra-subunit interactions in multi-domain assemblies. The isolated NTD cannot hydrolyze ATP at any detectable rate13,16, and we therefore applied the physiological nucleotide ATP instead of AMP-PNP. ACFs of reporter-containing NTDs recorded in absence and presence of ATP are shown in Figures 4b and 4c. ACFs of control samples lacking the engineered Trp showed a single decay on the ~2-ms time scale that resulted from translational diffusion of NTDs through the detection volume. Some residual fluctuations were observed in controls A112C and Q14C, which may arise from changes of polarity in the micro-environment of the label associated with conformational change. Reporters Q14C-A2W and A112C-S25W showed additional decays of substantial amplitude, which required a bi-exponential function to describe them accurately. These decays resulted from conformational fluctuations that were transformed into fluorescence fluctuations by PET. The major kinetic phase was on the ~400-µs scale. A second decay was on the ~7-µs scale but of negligible amplitude in comparison (Supplementary Table 4). To test if the ~400-µs phase truly arose from motion of the terminal β-strand, we applied the TrpZip motif to stabilize the β-strand on the domain (Figure 2a). Application of the TrpZip eliminated PET fluorescence fluctuations in the ACF, the decay of which was now well described by molecular diffusion only, showing that the β-strand was immobilized (Figure 4b). Results showed that N-terminal β-strand and lid are not rigid structures, despite their ordered appearance in crystallographic data, but are in fact highly mobile. We estimated the microscopic rate constants of unfolding of β-strand and lid from the observed amplitudes and time constants assuming a two-state equilibrium between fluorescent and fluorescence-quenched conformations (see Online Methods). The obtained rate constants were 1000±150 s-1 and 1500±70 s-1, respectively. Binding of ATP reduced the fluctuation amplitude of the lid significantly but had marginal effects on the β-strand (Figure 4b and 4c). Binding of ATP accelerated lid release to 2580±40 s-1, but the rate constant of β-strand release remained within error (810±100 s-1). We asked whether binding of ATP to the isolated NTD would induce full closure of the lid over the nucleotide-binding pocket. Fluorescence intensity time traces of reporter A110C-S51W showed that this was apparently not the case (Supplementary Figure 4a).
Next, we investigated the influence of the presence of the M-domain on dynamics of lid and β-strand in the NTD. We synthesized constructs that lacked the CTD (i.e. constructs consisting only of N- and M-domain separated by the charged linker; NM-domain), and that were therefore monomeric. NM-domains at low nM concentrations are not capable of hydrolyzing ATP13. ACFs of NM-domain constructs Q14C-A2W and A112C-S25W showed additional fluorescence fluctuations over those observed in control samples that lacked the engineered Trp, similar to the NTD, but the kinetics were more complex (Figure 4d and 4e). The main PET decay was an exponential on the 300-900 µs time scale. There were additional fluctuations of lower amplitude on the fast ns-µs scale that most likely resulted from transient intra-subunit interactions of N- and M-domains separated by the long and flexible charged linker (Supplementary Table 4). We could assign with confidence the 300-900 µs kinetic phases to motions of the β-strand and the lid because they were identified in the isolated NTD as single-exponential decays of similar time constants. Fits to the ACFs using a model that lacked an exponential phase on the 300-900 µs time scale did not describe the data well (Supplementary Figure 5). Such fits yielded artificially small diffusion time constants, which arose from an erroneous description of fluorescence fluctuations from diffusion and conformational changes on similar time scales, showing that conformational fluctuations on the 300-900 µs scale were truly present. From the observed amplitudes and time constants we estimated the rate constants of β-strand release and lid release to 540±130 s-1 and 1800±200 s-1, respectively (Figure 4f). Binding of ATP had no significant effect on the β-strand but doubled the rate constant of lid release to 3600±200 s-1, similar as observed for the NTD.
Finally, we investigated the influence of Aha1 on dynamics of β-strand and lid. Binding of Aha1 to the isolated NM-domain26,47 increased the diffusion time constant from 2.2 ms to 2.6 ms, which was consistent with the expected increase of molecular weight. The co-chaperone had weak effect on dynamics of the β-strand. However, it mobilized the lid significantly (Figure 4f). We found an increase of rate constant of lid release to 5800±800 s-1 upon binding of Aha1. ATP was not able to fully close the lid over the ATP-binding pocket in the NM-domain construct in complex with Aha1, which was evident from fluorescence intensity time traces of reporter S51C-A110W (Supplementary Figure 4b).
Discussion
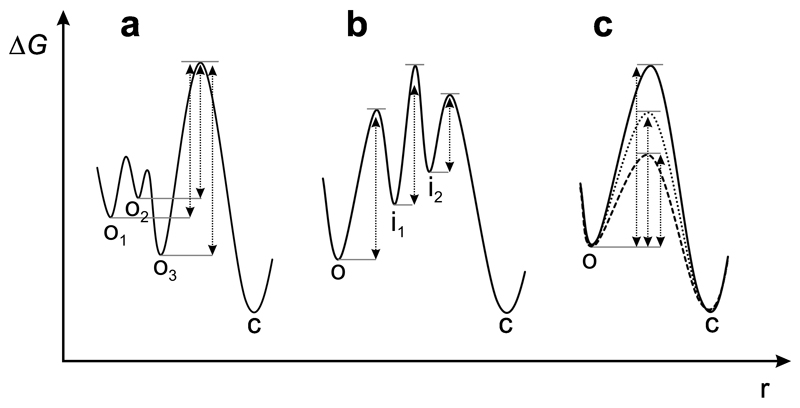
A range of structural studies show that binding of ATP to Hsp90 drives a set of local conformational switches that coordinate to the global process of domain rearrangements, referred to as N-terminal dimerization or closure of the molecular clamp, forming the ‘tense’ catalytically active conformation of the chaperone14,19–21,45. Previous FRET spectroscopy, which probes global domain rearrangements, shows multi-exponential kinetics of clamp-closure triggered by nucleotide binding. Multi-exponential kinetics was interpreted as reporting on formation of discrete intermediates along the conformational pathway23,24. For example, a slow exponential phase was assigned to slow closure of the lid as a first intermediate state in the catalytic cycle23. Multi-exponential kinetics in protein conformation, however, can have various origins in general, such as heterogeneity of the ground-state conformational ensemble from which transitions originate, population of discrete intermediates along the pathway of conformational change, or the presence of multiple pathways over free energy barriers of different height (Figure 5). Distinguishing these scenarios is not straightforward. For example, identification of intermediates along a folding pathway requires complex analysis of kinetic phases and their coupling48. Structural studies show that the apo state of Hsp90 is a heterogeneous ensemble of conformers that resemble beads on a string38,39. This ground-state conformational heterogeneity is thought to be responsible for the remarkable capacity of Hsp90 to accommodate structurally diverse clients49. Assuming ground-state conformational heterogeneity as the origin of multi-exponential kinetics in PET experiments, we calculated the mean rate constant of each conformational change from the sum of individual time constants weighted by the respective amplitudes. Obtained quantities were in good agreement with detected ATPase activities. It should be noted, however, that in vivo Hsp90 molecules progress repeatedly through the conformational cycle, which may change the relative populations of open-clamp conformations.
Figure 5. Possible origins of multi-exponential kinetics in protein dynamics.
Two-dimensional projection of a conformational free energy surfaces along an arbitrary reaction coordinate. (a) Ground-state heterogeneity. Multiple open-clamp conformations (o1-o3) of different free energy give rise to energy barriers of different height (broken arrows) along parallel pathways to the closed-clamp conformation (c). (b) On-pathway intermediates. Conformational change along a pathway containing a series of discrete intermediates (i1-i3) of different free energy. (c) Pathway heterogeneity. Open-clamp conformers of same free energy traverse to the closed-clamp conformation along different pathways that are characterized by different energy barrier heights.
Similarity of rate constants measured from remote sites suggests that conformational switches cooperate in formation of the catalytically active conformation. The interpretation was supported by mutagenesis experiments that showed similar modulation of kinetics of point mutants. Mutations that affected lid closure or docking of M- and N-domains, which dramatically impair ATPase activity, were strongly interdependent. We found weak coupling of β-strand swap with the other motions, suggesting an auxiliary role, possibly stabilizing the closed conformation of the dimer under high workload.
The high, inherent mobility of N-terminal β-strand and lid, detected by PET-FCS, has implications in the mechanisms of β-strand swap and lid closure, which are difficult to rationalize from static structures alone. The loose and dynamic β-strand can sense the temporarily vacant site on the neighboring subunit once the N-domains associate and then fold onto it. Despite the weak coupling we found of strand-swap with other motions, the N-terminus does play an important regulatory role in the chaperone. Metazoan Hsp90 homologs contain an extension of the N-terminus of 10-50 residues. These organisms have a substantially lower ATPase activity than yeast Hsp90. Deletion of this ‘strap’ in TRAP1 leads to acceleration of ATP hydrolysis, an effect that is also shown for a Δ8-mutant of yeast Hsp9037,50. N-terminal extensions in metazoan Hsp90s may have similar effects. The N-terminal β-strand might therefore provide a modulatory element where the energy barrier between open and closed states can be increased or reduced through addition or removal of stabilizing interactions on the parental subunit domain.
High mobility of the malleable lid primes the segment for rapid remodeling induced by binding of nucleotide, which was identified as the initial step of closure over the nucleotide-binding pocket. This initial event might release the self-association interface of the NTD early and prepare the lid for slow closure in cooperation with other motions.
The co-chaperone Aha1 is known to stimulate ATPase activity by remodeling the catalytic loop in the M-domain and by promoting an N-terminally closed state26,51. Aha1 consists of two-domains: The N-terminal domain (N-Aha1), which binds to the M-domain of Hsp9026, and the C-terminal domain (C-Aha1), which is thought to bind the nucleotide-bound form of Hsp90 where it stabilizes the N-terminally dimerized state52. N-Aha1 in isolation can stimulate ATPase activity to some extent while C-Aha1 cannot. Maximal stimulation of ATPase-activity requires binding of both domains of full-length Aha121,25,26,52. The main interaction occurs between N-Aha1 and the Hsp90 M-domain26, and is essentially independent of nucleotide25. N-Aha1 mediates constitutive association of Aha1 with Hsp90 and anchors C-Aha1 in close proximity to the Hsp90 N-domain. Here, we observed that Aha1 stabilized the N/M-associated state, in agreement with previous findings23,45, and accelerated all three local motions. It has been suggested that Aha1 helps “bypassing” a slowly formed closed-lid intermediate state of the NTD of Hsp9023. The proposal was amended recently by a new model suggesting that Aha1 initiates a partially closed lid and acts late on the nucleotide-bound N-terminally-dimerized conformation51. PET-FCS showed that Aha1 mobilizes the lid early in the apo-state of Hsp90. In support of this finding, NMR chemical shift perturbations show weak and transient interactions of C-Aha1 with the N-domain of Hsp9052. Lid-mutation T101I is re-activated by Aha145. Lid mobilization therefore emerges as an early mode of action of this co-chaperone.
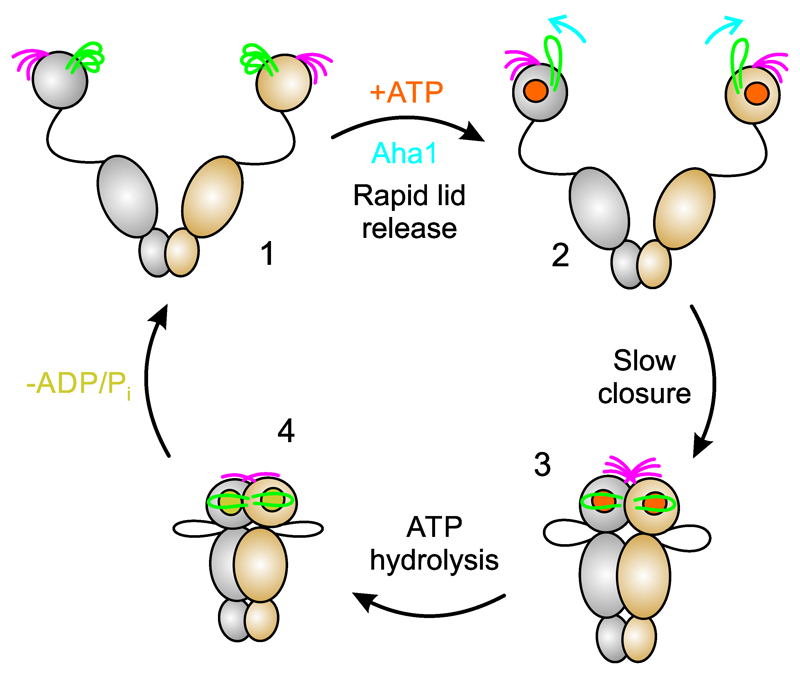
Figure 6 integrates our findings into the conformational cycle of Hsp90. At the beginning, Hsp90 is unconstrained and can adopt a plethora of flexible open-clamp conformations. Sub-populations of different free energy within a heterogeneous ensemble of conformers give rise to parallel pathways to the N-terminally dimerized, closed-clamp conformation, encountering different activation barriers. The lid in apo Hsp90 is not a static structure but dynamically populates an ensemble of conformers. Binding of ATP rapidly reconfigures the lid to an intermediate state, and this process likely releases the dimerization interface. Full closure of the lid over the nucleotide-binding pocket occurs slowly and in cooperation with inter- and intra-subunit association of N- and M-domains. The N-terminal β-strand of apo Hsp90 is highly mobile, which facilitates subunit swap upon association of N-domains. The co-chaperone Aha1 releases the lid early in the catalytic cycle, adding to the established modes of action, that is, modulation of the catalytic loop in the M-domain and stabilization of N/M-domain interactions26,52. Swap of the terminal β-strands, closure of the lid, and association of N- and M-domains cooperatively coordinate formation of the catalytically active conformation that hydrolyses ATP. Opening of the molecular clamp reconstitutes Hsp90 for the next catalytic cycle.
Figure 6. Integration of results into the chaperone catalytic cycle.
(1) At the beginning of the catalytic cycle, apo Hsp90 populates a heterogeneous ensemble of open-clamp conformers. Lid (green) and N-terminal β-strand (magenta) are highly mobile structural elements with sub-millisecond reconfiguration times. (2) Binding of ATP to the NTD leads to rapid release of the lid to an intermediate conformational state. The co-chaperone Aha1 pre-associates N- and M-domains but also remodels the lid segment for accelerated closure. (3) Closure of the molecular clamp involves cooperative action of conformational switches. Closure of the lid over the ATP-binding pocket, cross-subunit swap of β-strands, and association of the N- and M-domains are slow and interdependent. Swap of the terminal β-strands is weakly coupled with the other motions. (4) Hydrolysis of ATP leads to a compact, ADP-bound conformation, which relaxes to an open state with concomitant release of ADP and inorganic phosphate. Opening of the molecular clamp reconstitutes Hsp90 for the next catalytic cycle.
Online Methods
Protein synthesis, mutagenesis, and fluorescence modification
Engineered constructs for bacterial expression contained the genes of yeast Aha1, yeast Hsp82 in full-length, NTD (1-220) or NM-Domain (1-551) versions, with N-terminal His6-tag, cloned into a pRSET A vector (Invitrogen). Single-point mutants were generated using the QuikChange mutagenesis protocol (Stratagene). All constructs and mutants thereof were overexpressed in E. coli C41 (DE3) cells. His6-tagged proteins were isolated from bacterial cell lysate using Nickel-nitriloacetic acid chromatography. The eluate was loaded on an ion-exchange POROS® HQ column (Applied Biosystems) using 20 mM Tris-HCl, pH 8.0, as running buffer. Elution was performed applying a gradient from 0-1 M NaCl in 20 mM Tris-HCl, pH 8.0. In case of Cys mutants, 10 mM DTT was added to the protein solution prior to loading, and 1 mM DTT was added to running and elution buffer. Pooled fractions containing the protein were purified to homogeneity using size exclusion chromatography (SEC) on a Superdex 75 column (GE Healthcare), or, in case of full-length yeast Hsp90, on a Sephacryl™ S-400 column (GE Healthcare) equilibrated with buffer A (40 mM HEPES, pH 7.5, with the ionic strength adjusted to 200 mM using potassium chloride). In case of Cys mutants, SEC was performed using degassed buffer A. 10 mM DTT was added to the protein solution prior to SEC. Pooled fractions containing protein were concentrated using 10-kDa MWCO centrifugal concentrators (Vivaspin 20, Sartorius). Purity of synthesized proteins was confirmed by SDS-PAGE.
Single-point Cys mutants were fluorescently modified using the thiol-reactive maleimide derivative of the fluorophore AttoOxa11 (AttoTec). Labelling was carried out in buffer A that contained a 10-fold molar excess of tris(2-carboxyethyl)phosphine (TCEP) to prevent thiol oxidation. A 5-fold molar excess of dye and an incubating time of 2.5 hours at 25°C was applied. Labeled protein was isolated from excess dye using Sephadex G-25 resin (GE Healthcare) SEC.
ATPase assays
ATPase activities of Hsp90 constructs were measured using an enzyme-coupled ATPase assay as previously described15. A regenerating pyruvate kinase/lactate dehydrogenase (PK/LDH) linked assay, which is coupled to the oxidation of NADH to NAD+, was applied. Activity was measured as decrease of the NADH absorbance maximum at 340 nm in direct stoichiometry to ADP release. Assays were carried out at 25 ºC or 37 ºC in reaction buffer containing 0.2 mM NADH, 2 mM phosphoenol pyruvate, 50 U/ml pyruvate kinase, 50 U/ml lactate dehydrogenase, 2 mM ATP, 5 mM DTT, and 10 mM MgCl2 in buffer A. For co-chaperone experiments 20 µM Aha1 was added. Reactions were started by addition of Hsp90 at concentrations between 5-20 µM. The decrease in absorbance over time was detected using a V-650 spectrophotometer (Jasco). Background ATPase activity was recorded by inhibition of Hsp90 using geldanamycin (Cayman Chemical). The reaction buffer was prepared using 150 µM geldanamycin and 5-20 µM Hsp90. The reaction was started by addition of 2 mM ATP.
Time-resolved fluorescence experiments
Time-dependent fluorescence intensities were measured from Hsp90 samples in a quartz glass cuvette using a FP-6500 spectrofluorimeter (Jasco). Fluorescence was excited at 620 nm and emission intensities were recorded at a wavelength of 678 nm. Sample temperature was adjusted to 25 ºC using a peltier thermocouple. Hsp90 samples were prepared in buffer A containing 10 mM MgCl2 and 150 nM of AttoOxa11-labeled Hsp90 constructs. 5 µM non-labelled wild-type or mutant Hsp90 protein was added to ensure that only one subunit in hetero-dimeric constructs carried the fluorophore. Reactions were started by addition of 2 mM AMP-PNP or 4 mM ADP. Stock solutions of nucleotide were prepared by dissolving dry powder (≥93% purity; Sigma) in water to a concentration of 40 mM and stored at -80 ºC.
Stopped-flow fluorescence spectroscopy
Stopped-flow experiments were carried out on a SFM-300 machine (BioLogic Instruments) using a 639 nm diode laser as excitation source. The fluorescence signal was filtered using a long-pass optical filter (RazorEdge® 647RU, Semrock). 200 nM Oxa-labeled Hsp90 constructs were measured in solutions containing 5 µM wild-type Hsp90 or E162W and 20 µM Aha1 in buffer A containing 10 mM MgCl2. 4 mM AMP-PNP solution was added in a 1:1 mixing ratio such that the final concentration was 2 mM AMP-PNP. For measurement of rapid lid dynamics, samples contained 400 nM Oxa-labeled Hsp90 mutant S51W-A110C and 5 µM wild-type Hsp90. ADP was added in varying concentrations using stopped-flow syringes. Sample temperature was adjusted to 25 ºC using a circulating water bath.
PET-FCS experiments
PET-FCS was performed using a custom-built confocal fluorescence microscope setup described elsewhere53. Fluorescently modified Hsp90 constructs were diluted to 1 nM concentration in 50 mM phosphate buffer pH 7.5 containing 10 mM MgCl2 (with the solution ionic strength adjusted to 200 mM using potassium chloride). 0.3 mg/ml Protease-free bovine serum albumin and 0.05% Tween-20 were used as solution additives to suppress sample/glass-surface interactions. To study the influence of effector molecules, Hsp90 constructs were incubated with 20 µM Aha1 or 2 mM ATP prior to measurement. Samples were filtered through a 0.2 µm syringe filter, transferred onto a microscope slide, and covered by a cover slip. A 1-nM sample yielded an average of ~20 molecules in the detection focus of the microscope setup. Sample temperature was adjusted to 25 ºC using a custom-built objective heater. For each sample, three individual ACFs were recorded of 10 min measurement time each.
PET-FCS data analysis
ACFs were analyzed by fitting a model for globule diffusion in two dimensions and a sum of single-exponential relaxations:
| (1) |
τ is the lag time, N is the average number of molecules in the detection volume, τD is the experimental diffusion time constant, an and τn are the observed amplitude and time constant of the nth relaxation.
Rate constants of release of the lid and the N-terminal β-strand were calculated from kinetic quantities of the main PET decays in the ACFs (a1 and τ1; Supplementary Table 4) assuming a two-state transition between fluorescent and non-fluorescent conformational states. Validity of the assumption is supported by previous work on loop closure kinetics of unstructured model peptides measured using the same technique and analysis54, and by the fact that Oxa and Trp in aqueous solution form virtually non-fluorescent, π-π stacking complexes34. Microscopic rate constants of transitions to fluorescent and fluorescence-quenched states, kon and koff, respectively, were calculated from a1 and τ1:
| (2) |
| (3) |
In lid reporter A112C-S25W, the fully open-lid conformation is fluorescence-quenched (Figure 4a) and release of the lid leads to fluorescent conformation. The rate constant of lid release is thus kon. In β-strand reporter Q14C-A2W, the label is fluorescent as long as the β-strand is folded on the domain (Figure 4a) and gets fluorescence-quenched once it detaches. The rate constant of strand release is thus koff.
Supplementary Material
Supplementary Information is available.
Acknowledgments
The authors thank the Deutsche Forschungsgemeinschaft (grant # NE 1201/3-1 to H.N.) and the Wellcome Trust (Senior Investigator Award 095605/Z11/Z to L.H.P). A.S. was supported by a grant of the German Excellence Initiative to the Graduate School of Life Sciences (University of Würzburg).
Footnotes
Author contributions
A.S. designed experiments, synthesized modified protein material, performed rapid-mixing fluorescence experiments, performed ATPase assays, analyzed data, interpreted results, and wrote the paper. G.B. synthesized modified protein material, performed PET-FCS experiments, analyzed data, and interpreted results. D.A.H. synthesized modified protein material, performed PET-FCS experiments, and analyzed data. J.S. synthesized modified protein material, performed PET-FCS experiments, and analyzed data. L.H.P. interpreted results and wrote the paper. C.P. designed experiments, interpreted results, and wrote the paper. H.N. conceptually designed the research, designed experiments, analyzed data, interpreted results, and wrote the paper.
Competing financial interests:
The authors declare no competing financial interests.
References
- 1.Taipale M, Jarosz DF, Lindquist S. HSP90 at the hub of protein homeostasis: emerging mechanistic insights. Nat Rev Mol Cell Biol. 2010;11:515–528. doi: 10.1038/nrm2918. [DOI] [PubMed] [Google Scholar]
- 2.Saibil H. Chaperone machines for protein folding, unfolding and disaggregation. Nat Rev Mol Cell Biol. 2013;14:630–642. doi: 10.1038/nrm3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nagy PD, Wang RY, Pogany J, Hafren A, Makinen K. Emerging picture of host chaperone and cyclophilin roles in RNA virus replication. Virology. 2011;411:374–82. doi: 10.1016/j.virol.2010.12.061. [DOI] [PubMed] [Google Scholar]
- 4.Whitesell L, Lindquist SL. HSP90 and the chaperoning of cancer. Nat Rev Cancer. 2005;5:761–72. doi: 10.1038/nrc1716. [DOI] [PubMed] [Google Scholar]
- 5.Pearl LH, Prodromou C, Workman P. The Hsp90 molecular chaperone: an open and shut case for treatment. Biochem J. 2008;410:439–53. doi: 10.1042/BJ20071640. [DOI] [PubMed] [Google Scholar]
- 6.Trepel J, Mollapour M, Giaccone G, Neckers L. Targeting the dynamic HSP90 complex in cancer. Nat Rev Cancer. 2010;10:537–49. doi: 10.1038/nrc2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prodromou C, et al. Identification and structural characterization of the ATP/ADP-binding site in the Hsp90 molecular chaperone. Cell. 1997;90:65–75. doi: 10.1016/s0092-8674(00)80314-1. [DOI] [PubMed] [Google Scholar]
- 8.Jhaveri K, et al. Heat shock protein 90 inhibitors in the treatment of cancer: current status and future directions. Expert Opin Investig Drugs. 2014;23:611–28. doi: 10.1517/13543784.2014.902442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Neckers L, Workman P. Hsp90 molecular chaperone inhibitors: are we there yet? Clin Cancer Res. 2012;18:64–76. doi: 10.1158/1078-0432.CCR-11-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cullinan SB, Whitesell L. Heat shock protein 90: a unique chemotherapeutic target. Semin Oncol. 2006;33:457–65. doi: 10.1053/j.seminoncol.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 11.Meyer P, et al. Structural and functional analysis of the middle segment of hsp90: implications for ATP hydrolysis and client protein and cochaperone interactions. Mol Cell. 2003;11:647–58. doi: 10.1016/s1097-2765(03)00065-0. [DOI] [PubMed] [Google Scholar]
- 12.Vaughan CK, et al. Structure of an Hsp90-Cdc37-Cdk4 complex. Mol Cell. 2006;23:697–707. doi: 10.1016/j.molcel.2006.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Richter K, Muschler P, Hainzl O, Buchner J. Coordinated ATP Hydrolysis by the Hsp90 Dimer. J Biol Chem. 2001;276:33689–33696. doi: 10.1074/jbc.M103832200. [DOI] [PubMed] [Google Scholar]
- 14.Ali MM, et al. Crystal structure of an Hsp90-nucleotide-p23/Sba1 closed chaperone complex. Nature. 2006;440:1013–7. doi: 10.1038/nature04716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Panaretou B, et al. ATP binding and hydrolysis are essential to the function of the Hsp90 molecular chaperone in vivo. EMBO J. 1998;17:4829–36. doi: 10.1093/emboj/17.16.4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prodromou C, et al. The ATPase cycle of Hsp90 drives a molecular 'clamp' via transient dimerization of the N-terminal domains. EMBO J. 2000;19:4383–92. doi: 10.1093/emboj/19.16.4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maloney A, et al. Gene and protein expression profiling of human ovarian cancer cells treated with the heat shock protein 90 inhibitor 17-allylamino-17-demethoxygeldanamycin. Cancer Res. 2007;67:3239–53. doi: 10.1158/0008-5472.CAN-06-2968. [DOI] [PubMed] [Google Scholar]
- 18.Roe SM, et al. Structural basis for inhibition of the Hsp90 molecular chaperone by the antitumor antibiotics radicicol and geldanamycin. J Med Chem. 1999;42:260–6. doi: 10.1021/jm980403y. [DOI] [PubMed] [Google Scholar]
- 19.Shiau AK, Harris SF, Southworth DR, Agard DA. Structural Analysis of E. coli hsp90 Reveals Dramatic Nucleotide-Dependent Conformational Rearrangements. Cell. 2006;127:329–340. doi: 10.1016/j.cell.2006.09.027. [DOI] [PubMed] [Google Scholar]
- 20.Pearl LH, Prodromou C. Structure and mechanism of the Hsp90 molecular chaperone machinery. Annu Rev Biochem. 2006;75:271–94. doi: 10.1146/annurev.biochem.75.103004.142738. [DOI] [PubMed] [Google Scholar]
- 21.Prodromou C. The 'active life' of Hsp90 complexes. Biochim Biophys Acta. 2012;1823:614–23. doi: 10.1016/j.bbamcr.2011.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Graf C, Stankiewicz M, Kramer G, Mayer MP. Spatially and kinetically resolved changes in the conformational dynamics of the Hsp90 chaperone machine. EMBO J. 2009;28:602–13. doi: 10.1038/emboj.2008.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hessling M, Richter K, Buchner J. Dissection of the ATP-induced conformational cycle of the molecular chaperone Hsp90. Nat Struct Mol Biol. 2009;16:287–293. doi: 10.1038/nsmb.1565. [DOI] [PubMed] [Google Scholar]
- 24.Mickler M, Hessling M, Ratzke C, Buchner J, Hugel T. The large conformational changes of Hsp90 are only weakly coupled to ATP hydrolysis. Nat Struct Mol Biol. 2009;16:281–6. doi: 10.1038/nsmb.1557. [DOI] [PubMed] [Google Scholar]
- 25.Panaretou B, et al. Activation of the ATPase activity of hsp90 by the stress-regulated cochaperone aha1. Mol Cell. 2002;10:1307–18. doi: 10.1016/s1097-2765(02)00785-2. [DOI] [PubMed] [Google Scholar]
- 26.Meyer P, et al. Structural basis for recruitment of the ATPase activator Aha1 to the Hsp90 chaperone machinery. EMBO J. 2004;23:511–9. doi: 10.1038/sj.emboj.7600060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roe SM, et al. The Mechanism of Hsp90 regulation by the protein kinase-specific cochaperone p50(cdc37) Cell. 2004;116:87–98. doi: 10.1016/s0092-8674(03)01027-4. [DOI] [PubMed] [Google Scholar]
- 28.Neuweiler H, Sauer M. Using photoinduced charge transfer reactions to study conformational dynamics of biopolymers at the single-molecule level. Curr Pharm Biotechnol. 2004;5:285–98. doi: 10.2174/1389201043376896. [DOI] [PubMed] [Google Scholar]
- 29.Doose S, Neuweiler H, Sauer M. Fluorescence quenching by photoinduced electron transfer: a reporter for conformational dynamics of macromolecules. ChemPhysChem. 2009;10:1389–98. doi: 10.1002/cphc.200900238. [DOI] [PubMed] [Google Scholar]
- 30.Neuweiler H, Johnson CM, Fersht AR. Direct observation of ultrafast folding and denatured state dynamics in single protein molecules. Proc Natl Acad Sci U S A. 2009;106:18569–18574. doi: 10.1073/pnas.0910860106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Neuweiler H, Banachewicz W, Fersht AR. Kinetics of chain motions within a protein-folding intermediate. Proc Natl Acad Sci U S A. 2010;107:22106–10. doi: 10.1073/pnas.1011666107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sauer M, Neuweiler H. PET-FCS: probing rapid structural fluctuations of proteins and nucleic acids by single-molecule fluorescence quenching. Methods Mol Biol. 2014;1076:597–615. doi: 10.1007/978-1-62703-649-8_27. [DOI] [PubMed] [Google Scholar]
- 33.Schuler B, Hofmann H. Single-molecule spectroscopy of protein folding dynamics-expanding scope and timescales. Curr Opin Struct Biol. 2013;23:36–47. doi: 10.1016/j.sbi.2012.10.008. [DOI] [PubMed] [Google Scholar]
- 34.Vaiana AC, et al. Fluorescence quenching of dyes by tryptophan: interactions at atomic detail from combination of experiment and computer simulation. J Am Chem Soc. 2003;125:14564–72. doi: 10.1021/ja036082j. [DOI] [PubMed] [Google Scholar]
- 35.Mishra P, Bolon DN. Designed Hsp90 heterodimers reveal an asymmetric ATPase-driven mechanism in vivo. Mol Cell. 2014;53:344–50. doi: 10.1016/j.molcel.2013.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roughley SD, Hubbard RE. How well can fragments explore accessed chemical space? A case study from heat shock protein 90. J Med Chem. 2011;54:3989–4005. doi: 10.1021/jm200350g. [DOI] [PubMed] [Google Scholar]
- 37.Richter K, Reinstein J, Buchner J. N-terminal residues regulate the catalytic efficiency of the Hsp90 ATPase cycle. J Biol Chem. 2002;277:44905–44910. doi: 10.1074/jbc.M208457200. [DOI] [PubMed] [Google Scholar]
- 38.Krukenberg KA, Forster F, Rice LM, Sali A, Agard DA. Multiple conformations of E. coli Hsp90 in solution: insights into the conformational dynamics of Hsp90. Structure. 2008;16:755–65. doi: 10.1016/j.str.2008.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Southworth DR, Agard DA. Species-dependent ensembles of conserved conformational states define the Hsp90 chaperone ATPase cycle. Mol Cell. 2008;32:631–40. doi: 10.1016/j.molcel.2008.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cochran AG, Skelton NJ, Starovasnik MA. Tryptophan zippers: stable, monomeric beta-hairpins. Proc Natl Acad Sci U S A. 2001;98:5578–83. doi: 10.1073/pnas.091100898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Santiveri CM, Jimenez MA. Tryptophan residues: scarce in proteins but strong stabilizers of beta-hairpin peptides. Biopolymers. 2010;94:779–90. doi: 10.1002/bip.21436. [DOI] [PubMed] [Google Scholar]
- 42.Cunningham CN, Southworth DR, Krukenberg KA, Agard DA. The conserved arginine 380 of Hsp90 is not a catalytic residue, but stabilizes the closed conformation required for ATP hydrolysis. Protein Sci. 2012;21:1162–1171. doi: 10.1002/pro.2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McLaughlin SH, Ventouras LA, Lobbezoo B, Jackson SE. Independent ATPase activity of Hsp90 subunits creates a flexible assembly platform. J Mol Biol. 2004;344:813–826. doi: 10.1016/j.jmb.2004.09.055. [DOI] [PubMed] [Google Scholar]
- 44.Wegele H, Muschler P, Bunck M, Reinstein J, Buchner J. Dissection of the contribution of individual domains to the ATPase mechanism of Hsp90. J Biol Chem. 2003;278:39303–39310. doi: 10.1074/jbc.M305751200. [DOI] [PubMed] [Google Scholar]
- 45.Siligardi G, et al. Co-chaperone regulation of conformational switching in the Hsp90 ATPase cycle. J Biol Chem. 2004;279:51989–98. doi: 10.1074/jbc.M410562200. [DOI] [PubMed] [Google Scholar]
- 46.Prodromou C, Pearl LH. Structure and functional relationships of Hsp90. Curr Cancer Drug Targets. 2003;3:301–23. doi: 10.2174/1568009033481877. [DOI] [PubMed] [Google Scholar]
- 47.Koulov AV, et al. Biological and structural basis for Aha1 regulation of Hsp90 ATPase activity in maintaining proteostasis in the human disease cystic fibrosis. Mol Biol Cell. 2010;21:871–84. doi: 10.1091/mbc.E09-12-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gianni S, Ivarsson Y, Jemth P, Brunori M, Travaglini-Allocatelli C. Identification and characterization of protein folding intermediates. Biophys Chem. 2007;128:105–13. doi: 10.1016/j.bpc.2007.04.008. [DOI] [PubMed] [Google Scholar]
- 49.Krukenberg KA, Street TO, Lavery LA, Agard DA. Conformational dynamics of the molecular chaperone Hsp90. Q Rev Biophys. 2011;44:229–255. doi: 10.1017/S0033583510000314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lavery LA, et al. Structural Asymmetry in the Closed State of Mitochondrial Hsp90 (TRAP1) Supports a Two-Step ATP Hydrolysis Mechanism. Mol Cell. 2014;53:330–343. doi: 10.1016/j.molcel.2013.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li J, Richter K, Reinstein J, Buchner J. Integration of the accelerator Aha1 in the Hsp90 co-chaperone cycle. Nat Struct Mol Biol. 2013;20:326–31. doi: 10.1038/nsmb.2502. [DOI] [PubMed] [Google Scholar]
- 52.Retzlaff M, et al. Asymmetric activation of the hsp90 dimer by its cochaperone aha1. Mol Cell. 2010;37:344–54. doi: 10.1016/j.molcel.2010.01.006. [DOI] [PubMed] [Google Scholar]
- 53.Ries J, Schwarze S, Johnson CM, Neuweiler H. Microsecond folding and domain motions of a spider silk protein structural switch. J Am Chem Soc. 2014;136:17136–44. doi: 10.1021/ja508760a. [DOI] [PubMed] [Google Scholar]
- 54.Daidone I, Neuweiler H, Doose S, Sauer M, Smith JC. Hydrogen-bond driven loop-closure kinetics in unfolded polypeptide chains. PLoS Comput Biol. 2010;6:e1000645. doi: 10.1371/journal.pcbi.1000645. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.