Abstract
Little is known about the immunoediting process in precancerous lesions. We explored this aspect of benign colorectal adenomas with a descriptive analysis of the immune pathways and immune cells whose regulation is linked to the morphology and size of these lesions. Two series of polypoid and nonpolypoid colorectal adenomas were used in this study: 1) 84 samples (42 lesions, each with matched samples of normal mucosa) whose gene expression data were used to quantify the tumor morphology- and size-related dysregulation of immune pathways collected in the Molecular Signature Database, using Gene Set Enrichment Analysis; 2) 40 other lesions examined with immunohistochemistry to quantify the presence of immune cells in the stromal compartment. In the analysis of transcriptomic data, 429 immune pathways displayed significant differential regulation in neoplasms of different morphology and size. Most pathways were significantly upregulated or downregulated in polypoid lesions versus nonpolypoid lesions (regardless of size). Differential pathway regulation associated with lesion size was observed only in polypoid neoplasms. These findings were mirrored by tissue immunostaining with CD4, CD8, FOXP3, MHC-I, CD68, and CD163 antibodies: stromal immune cell counts (mainly T lymphocytes and macrophages) were significantly higher in polypoid lesions. Certain markers displayed significant size-related differences regardless of lesion morphology. Multivariate analysis of variance showed that the marker panel clearly discriminated between precancerous lesions of different morphologies and sizes. Statistical analysis of immunostained cell counts fully support the results of the transcriptomic data analysis: the density of infiltration of most immune cells in the stroma of polypoid precancerous lesions was significantly higher than that observed in nonpolypoid lesions. Large neoplasms also have more immune cells in their stroma than small lesions. Immunoediting in precancerous colorectal tumors may vary with lesion morphology and stage of development, and this variability could influence a given lesion’s trajectory to cancer.
Introduction
The immune system plays a Janus-like role in the development and progression of cancer, exerting tumor-promoting and tumor-suppressive effects. This duality is the basis of the process known as immunoediting, whereby the immune system shapes (or edits) the evolution of tumorigenesis, qualitatively and quantitatively [1]. Outcomes envisioned by this model range from 1) the outright elimination of tiny, nascent neoplastic lesions by joint intervention of the innate and adaptive immune systems; 2) a state of equilibrium between tumor cells and host, during which the adaptive immune system checks frank outgrowth and maintains a state mildly conducive to slow neoplastic proliferation; and 3) tumor escape, characterized by the emergence, under selective immune pressure, of decreasingly immunogenic neoplastic cell populations and a state of frank immunosuppression [1]. Recognition and ongoing characterization of these phenomena have prompted attempts to combat or control cancer by pharmaceutical modulation of the immunoediting process (e.g., enhancing the presence of immune effector cells with anti-tumor activities, inhibiting or eliminating molecular and cellular mediators of cancer-induced immunosuppression). The mechanisms underlying the equilibrium phase are of particular interest. This state of relative dormancy is typical of the benign, preinvasive stages of tumorigenesis, and it can keep malignancy at bay for years—up to two decades in some cases of colorectal neoplasia [2, 3, 4]. Prolongation or durable recapitulation of this state is thus regarded as a potentially achievable end point of immunotherapy [1]. The incidence of precancerous colorectal lesions and/or of “interval” cancers (those occurring before the follow-up colonoscopy) can be reduced by chemopreventive interventions with aspirin or other nonsteroidal anti-inflammatory drugs, which alter the tumor microenvironment, rendering it more conducive to the elimination of nascent tumors or the arrest or suppression of the growth of larger lesions [5, 6].
Little is known about the mechanisms underpinning the equilibrium phase in colorectal tumorigenesis. The immune landscape of epithelial tumors is highly complex, and the characteristics (cell types, extension, functionality) of intralesional immune-cell populations vary with the cell-type origin of the tumor and the site where it develops. In large bowel neoplasms, for example, the superficial layer of epithelial cells is the only barrier between the tumor stroma and the bacterial flora of the gut. The composition of the immune infiltrate in these tumors is thus likely to reflect its dual nature, as a response to immunogenic epithelial tumor-cell antigens and to any microbial products that pass through the neoplastic epithelium.
Immune cell infiltrates in invasive colorectal cancers have been well-characterized [7–21], but less is known about the immunomes of precancerous colorectal tumors [22–28]. The present study was an attempt to close that gap.
Precancerous colorectal lesions vary widely in morphology, size, and histology. Their gross appearance at endoscopy is broadly classified as “polypoid” or “nonpolypoid” [29]. The former tumors protrude into the gut lumen and are attached to the mucosa with a pedicle or stalk (pedunculated lesions, type Ip) or with a shorter, broader base (sessile lesions, type Is). Nonpolypoid lesions are still erroneously described as “flat” by many authors, although most are actually slightly elevated (< 2.5 mm above the mucosal surface; type IIa). Those that are truly flat or slightly depressed are rare (reviewed in [4]). In both morphological classes, increasing size reflect the lesion’s advancement on the road to cancer [30]. Most precancerous colorectal lesions—polypoid or nonpolypoid—are adenomas, and their degree of dysplasia increases with their size. Nonadenomatous precancerous lesions, the so-called sessile serrated adenomas/polyps, are much less common and rarely display epithelial dysplasia [31].
For years, the prevalence of nonpolypoid colorectal neoplasms in Western countries has been markedly underestimated. Today, however, thanks to improved awareness and training of colonoscopists and to advances in endoscopic technology, these lesions represent up to ~40% of the precancerous lesions identified during colonoscopy, particularly those found in the proximal colon [32, 33]. Some investigators have suggested that these tumors progress more rapidly to cancer than polypoid lesions [34], but in other studies the frequencies of carcinoma in nonpolypoid lesions resembled [35,36] or was lower than [33] that observed in polypoid lesions. The discrepancies are largely due to inconsistencies between studies in the use of endoscopic and histologic classifications of precancerous colorectal tumors (reviewed in [37]). At the molecular level (i.e., gene expression, genetic and epigenetic alterations), however, unequivocal differences have been documented between the epithelia of nonpolypoid and polypoid lesions [37, 38]. Therefore, these two categories of lesions can reasonably be expected to differ in terms of their tumor microenvironments, immune-cell infiltrates in particular.
In this study, we used gene set enrichment analysis (GSEA) [39] to explore a gene expression data set derived from endoscopic biopsies of polypoid and nonpolypoid colorectal adenomas. Our aim was to identify immune pathway regulation profiles related to lesion morphology and/or size. The results were validated by immunohistochemical analysis of formalin-fixed, paraffin-embedded (FFPE) sections from a second series of precancerous colorectal tissues.
Results
Gene set enrichment analysis
To identify biological pathways that might play key roles in the development of precancerous lesions in the colorectum, we used gene set enrichment analysis (GSEA) (Subramanian et al. 2005) with the Molecular Signatures Database (MSigDB) to explore the transcriptomic profiles of 42 precancerous colorectal neoplasms: 17 were polypoid lesions (9 small, 8 large), and 25 were nonpolypoid (10 small, 15 large) (see Methods for details). The original data had been obtained with microarray analysis of endoscopic mucosal biopsies containing neoplastic epithelium as well as stroma. The results were normalized to matched samples of non-neoplastic colorectal mucosa.
GSEA of our transcriptomic data using all the collections in the MSigDB revealed enrichment for immune-related pathways in the set of genes differentially expressed in the two morphologic classes of lesions. We therefore restricted our analysis to the C7 collection, which included 1910 gene sets representing various cell states and perturbations within the immune system. Five group comparisons were performed: 1. polypoid vs. nonpolypoid (all lesion sizes); 2. polypoid vs. nonpolypoid (small lesions only); 3. polypoid vs. nonpolypoid (large lesions only); 4. small vs. large (polypoid lesions only); 5. small vs. large (nonpolypoid lesions only). Diameter, the standard measurement of lesion size in endoscopic reports, was analyzed as an index of the precancerous lesions’ malignant potential [30].
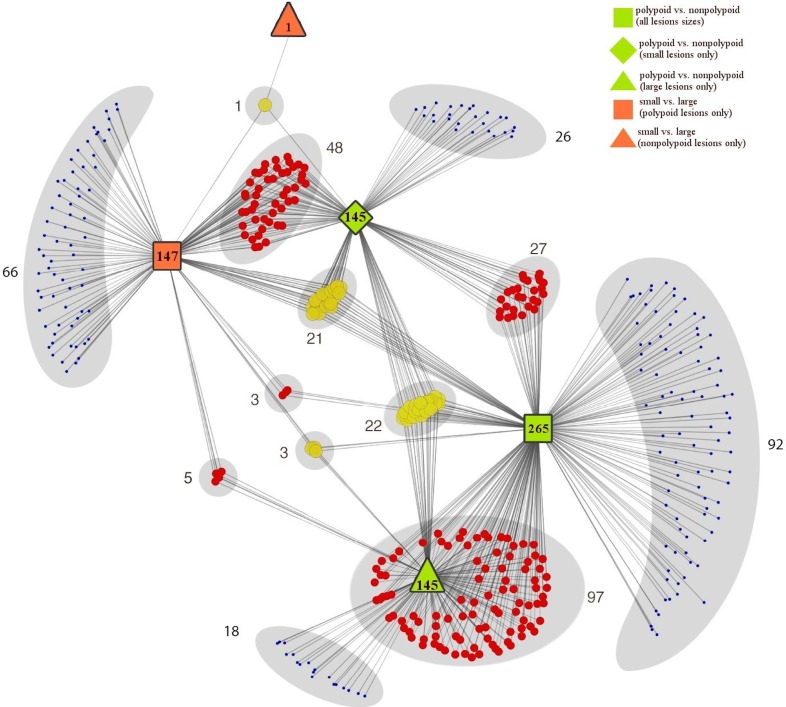
Fig 1and S1 Table show the 429 immune pathways that displayed significant differential regulation (p-values < 0.05) in the GSEA of one or more of the five group comparisons. Two hundred sixty-five pathways were significantly upregulated (n = 219) or downregulated (n = 46) in polypoid lesions relative to their nonpolypoid counterparts (all sizes). This morphology-associated difference was equally striking within both size classes: 145 immune pathways were upregulated (n = 35) or downregulated (n = 110) in small polypoid lesions compared with small nonpolypoid lesions, and the same number were differentially regulated (144 upregulations, 1 downregulation) in large polypoid lesions relative to large lesions with nonpolypoid morphologies. Lesion size was also associated with differential regulation of numerous immune pathways in the polypoid lesions (4 upregulated pathways and 143 downregulated pathways in small vs. large polypoid tumors). In the nonpolypoid group, only 1 pathway displayed downregulation in small lesions compared with the large lesions. Collectively, these findings strongly suggest that the immunologic signatures of polypoid adenomas are markedly different from those of their nonpolypoid counterparts, independently of their size. However, the signature of polypoid lesions also appears to change appreciably with lesion size.
Fig 1. Distribution of immune pathways displaying significant dysregulation in GSEA analysis of the five group comparisons.
Colored circles represent 429 pathways that exhibited significant dysregulation (p-value < 0.05) in the GSEA analysis of one (blue), two (red), or three (yellow) of the five group comparisons: 1) polypoid vs. nonpolypoid (all lesions sizes, green squares); 2) polypoid vs. nonpolypoid (small lesions only, green diamonds); 3) polypoid vs. nonpolypoid (large lesions only, green triangles.); 4) small vs. large (polypoid lesions only, orange squares.); 5) small vs. large (nonpolypoid lesions only, orange triangle). The complete list of differentially regulated pathways with their p-values is reported in S1 Table.
Immunohistochemistry studies
The results reported above were based on transcriptomic analysis of composite tissue biopsies with epithelial to stromal cell ratios of around 2:1. Assuming they are valid, one would expect to see similar trends emerge from analyses restricted to the stromal immune-cell landscape of precancerous colorectal lesions. We therefore examined a second, archival series of 40 colorectal adenomas (see Methods for details)—19 polypoid lesions (10 small, 9 large), 21 nonpolypoid lesions (10 small, 11 large). Immunohistochemistry was used for this purpose since it preserves tissue architecture and allows quantification of cellularity in specific tumor regions. These lesions were subjected to the same five group comparisons used for the pathway analysis.
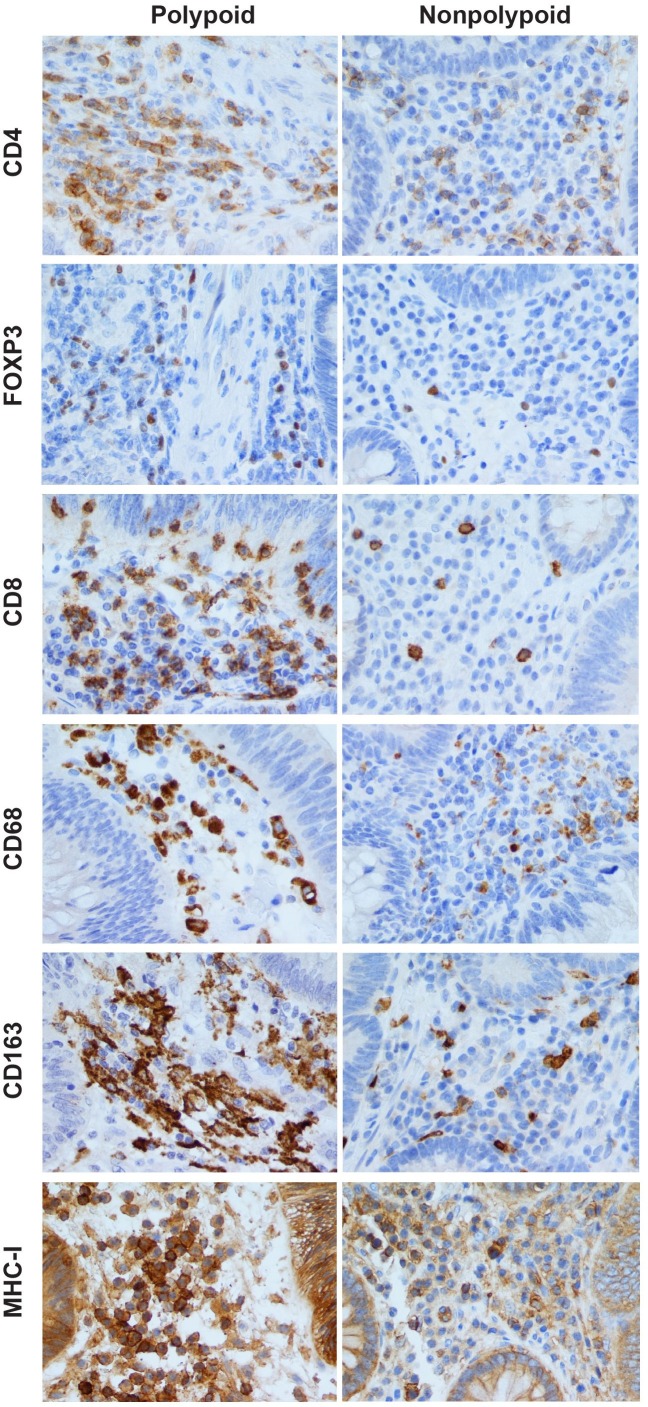
The immune cell populations most highly represented in the pathways shown in S1 Table were T-lymphocytes and macrophages (~50% and ~6% of the pathways, respectively); B-cell, NK-cell, and other immune cell signatures were all substantially less common. Serial sections of FFPE tissues were thus stained with antibodies against the T-lymphocyte markers CD4, CD8, and FOXP3 and the macrophage markers CD68 and CD163 (see Methods). Antibody against a sixth marker, MHC-I, was used to obtain a general, nonspecific count of stromal cells, including certain immune cells that were not covered by the other markers we used (e.g., dendritic cells, natural killer cells) (see Discussion). Fig 2shows representative findings for each marker in the stroma of polypoid and nonpolypoid colorectal adenomas.
Fig 2. Immunohistochemical assessment of stromal immune cell infiltrates in polypoid and nonpolypoid colorectal adenomas.
CD4+, FOXP3+, CD8+, CD68+, CD163+, and MHC-I+ immune cell densities (i.e., number of positively-stained cells per unit area; see Methods) in the stroma of polypoid adenomas were consistently higher than those observed in nonpolypoid lesions. Intraepithelial CD8+ (cytotoxic) T cells are also seen in the epithelial compartments of both types of tumor. In calculating CD68+ and CD163+ macrophage densities, we counted only nuclei surrounded by CD68+ or CD163+ granulations and excluded elongated or fragmented cells with granular cytoplasmic positivity. MHC-I labelling (granular, compact, or membranous) was observed in a variety of stromal cells. MHC-I+ epithelial cells were also present in both types of tumor, but their presence was not quantified in this study. Magnification for all panels: 400X.
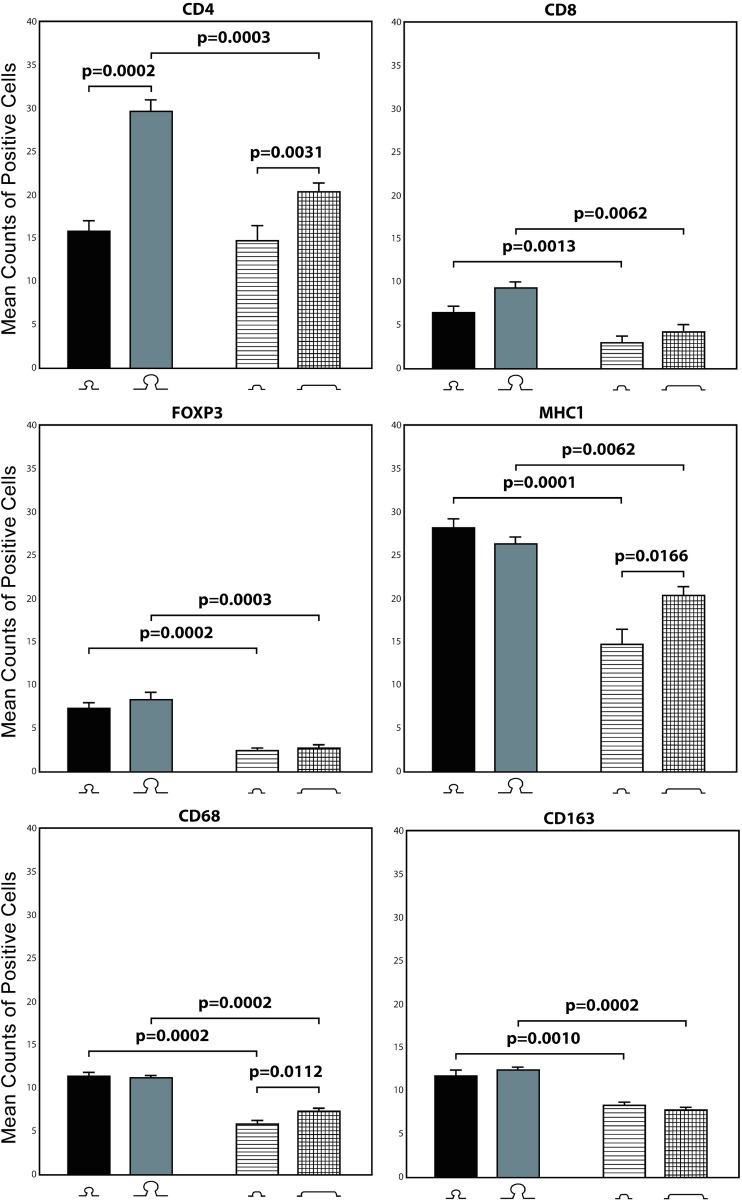
As shown in Fig 3and Table 1, for each marker the number of positively labelled stromal immune cells was significantly higher in polypoid than nonpolypoid lesions. These differences were generally independent of lesion size, i.e., the increased presence in polypoid adenomas was still significant when analysis was confined exclusively to the subset of small or large tumors. CD4+ cells were the exception: the higher cell counts in polypoid lesions vs. nonpolypoid were significant only in large lesions. As for size-related differences, CD4+ cell counts were also significantly higher in large polypoid lesions than in small lesions with the same morphology. In nonpolypoid lesions, CD4+, MHC-I+, and CD68+ cells were significantly more common in those that were large. (CD8+ T cells were also present within the neoplastic epithelium, but their numbers in this compartment showed no significant morphology- or size-related variations, and they were thus excluded from the statistical analysis.)
Fig 3. Immunohistochemical morphometric analysis of stromal immune cells.
Bars represent mean (SE) CD4+, CD8+, FOXP3+, MHC-I+, CD68+ and CD163+ cell counts for the following groups (from left to right): small polypoid lesions, large polypoid lesions, small nonpolypoid lesions, large nonpolypoid lesions. Horizontal bars with P values indicate significant differences between groups (Mann-Whitney test) (see Table 1for details).
Table 1. Significant differences between mean labelled immune cell counts in the five group comparisons.
| Group Comparisons | Group with higher counts | CD4+ | CD8+ | FOXP3+ | MHC-I+ | CD68+ | CD163+ |
|---|---|---|---|---|---|---|---|
| polypoid vs. nonpolypoid (all sizes) | polypoid | 10^-5 | 10^-7 | 10^-6 | 10^-8 | 10^-7 | |
| polypoid vs. nonpolypoid (small lesions only) | polypoid | 0.0013 | 0.0002 | 0.0001 | 0.0002 | 0.001 | |
| polypoid vs. nonpolypoid (large lesions only) | polypoid | 0.0003 | 0.0062 | 0.0003 | 0.0062 | 0.0002 | 0.0002 |
| small vs. large (polypoid lesions only) | large | 0.0002 | |||||
| small vs. large (nonpolypoid lesions only) | large | 0.0031 | 0.0166 | 0.0112 |
P-values, (Mann-Whitney test) are shown for each difference; statistical significance was set at p-value < 0.05. The direction of mean counts difference is shown for each marker in the five group comparisons.
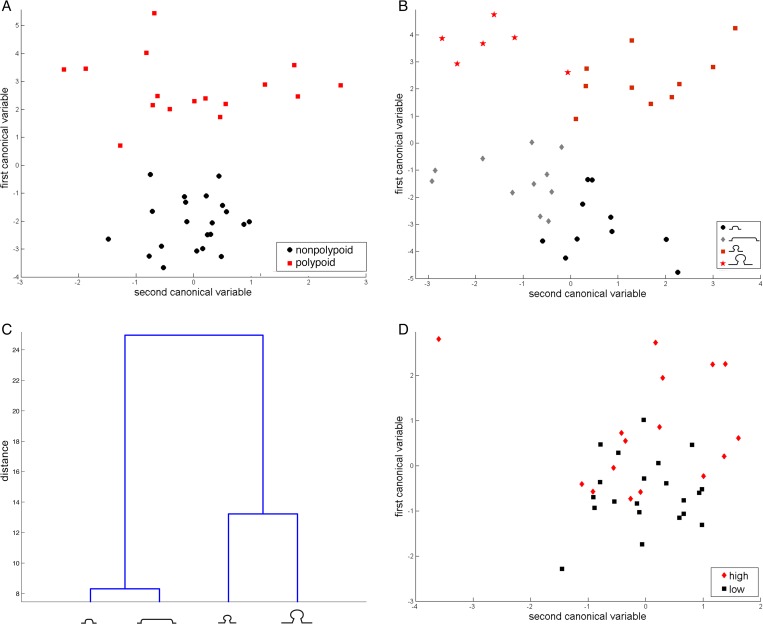
One-way multivariate analysis of variance (MANOVA) was then used to compare the multivariate means of the lesion groups in the five group comparisons (Table 2). Significant p-values were found in all five group comparisons, indicating that the number of cells labelled with the 6 immune markers we used effectively discriminated between adenomas of different shapes and sizes. When only lesion morphology was considered, the canonical analysis by MANOVA revealed that polypoid lesions were perfectly separated from nonpolypoid lesions along the axis of the first canonical variable (Fig 4A). When both morphology and size were considered, small and large lesions were clearly separated along the axis of the second canonical variable in both morphological classes (Fig 4B). Indeed, the main intergroup differences were related to lesion morphology and, within each morphology class, to lesion size (Fig 4C).
Table 2. Multivariate analysis of the immune cell counts in the five group comparisons.
| Comparison | p-value |
|---|---|
| polypoid vs. nonpolypoid (all sizes) | 10−11 |
| polypoid vs. nonpolypoid (small lesions only) | 10−5 |
| polypoid vs. nonpolypoid (large lesions only) | 10−5 |
| small vs. large (polypoid lesions only) | 0.001 |
| small vs. large (nonpolypoid lesions only) | 10−4 |
P-values were computed by MANOVA test.
Fig 4. Multivariate analysis of polypoid and nonpolypoid lesions (small and/or large) using MANOVA test.
A) Scatter plot of the first two canonical variables for all polypoid (red squares) and all nonpolypoid lesions (black circles); B) scatter plot of the first two canonical variables for small polypoid lesions (red squares), large polypoid lesions (red stars), small nonpolypoid lesions (black circles), large nonpolypoid lesions (gray diamonds); C) dendogram plot of group mean clusters; D) scatter plot of the first two canonical variables for all adenomas with high-grade dysplasia (red diamonds) and all adenomas with low-grade dysplasia (black squares), regardless of morphology and size. In the scatter plots, each dot represents one tissue sample.
Because lesion size is positively correlated with the degree of epithelial cell dysplasia in both polypoid and nonpolypoid adenomas [30], we analyzed the possible association between adenoma diameters and stromal immune cell counts. Because the number of samples was small, we compared lesions with high and low degrees of dysplasia only in the following groups of lesions: 1) all 39 lesions (one small, nonpolypoid lesion was excluded because it displayed histologic features of a sessile serrated adenoma/polyp, see Methods); 2) the 19 polypoid lesions; 3) the 20 nonpolypoid lesions; 4) the 19 small (polypoid and nonpolypoid) lesions; 5) the 20 large (polypoid and nonpolypoid) lesions (Table 3). The degree of epithelial dysplasia was significantly associated only with stromal counts of CD4+ and CD8+ cells. They were significantly more numerous in lesions displaying high-grade dysplasia (p-values of 0.002 and 0.048, respectively), regardless of morphology or size. In the polypoid lesion group, tumors of all sizes with high-grade dysplasia had significantly higher CD4+ cell counts (p-value = 0.003 vs. polypoid lesions with low-grade dysplasia). In the small lesion subset, tumors with high-grade dysplasia had significantly higher CD8+ cell counts (p-value = 0.026 vs. small lesions with low-degree dysplasia). The difference between tumors with high- and low-grade dysplasia displayed borderline significance in the group composed of all 39 lesions (p-value = 0.049), while the differences observed in the remaining four groups were not statistically significant. This finding suggests that the 6-marker panel does not discriminate well between lesions with high- and low-grade dysplasia. Indeed, scatter plots of these data produced two overlapping clouds of points (Fig 4D).
Table 3. Significant increases in labelled immune cell counts in lesions with high- vs. low-grade dysplasia.
| Comparison | group | CD4 | CD8 | FOXP3 | MHC-I | CD68 | CD163 |
|---|---|---|---|---|---|---|---|
| high- vs. low-grade dysplasia | all lesions | 0.002 | 0.048 | - | - | - | - |
| high- vs. low-grade dysplasia | polypoid | 0.003 | - | - | - | - | - |
| high- vs. low-grade dysplasia | nonpolypoid | - | - | - | - | - | - |
| high- vs. low-grade dysplasia | small | - | 0.026 | - | - | - | - |
| high- vs. low-grade dysplasia | large | - | - | - | - | - | - |
p-values computed by Mann-Whitney test are shown, and the criteria for statistical significance was set at p-value < 0.05.
Discussion
Our GSEA of the transcriptomes of precancerous colorectal lesions revealed major differences between the immune signatures of tumors with polypoid and nonpolypoid morphologies. Within the polypoid group, significant differences also emerged between small and large lesions. The most striking differences (S1 Table) were the focus of our stromal immunohistochemistry studies in a second set of colorectal adenomas. The latter analyses confirmed that T lymphocyte and macrophage cell counts were significantly higher in polypoid lesions (versus nonpolypoid tumors), regardless of size. Significant size-associated differences were also observed for certain immune markers in polypoid or nonpolypoid lesions (Table 1). Indeed, the panel clearly discriminated between precancerous lesions of different shapes and sizes (Table 2and Fig 4). When the variable epithelial cell dysplasia was included in the analysis, statistics were applicable to groups with a reasonable number of lesions. The positive association between high-grade dysplasia and immune-cell density in the stroma of adenomas (Table 3and Fig 4) displayed only borderline significance and thus requires further analysis in a larger series of tissues.
Ours is the first attempt to determine how the composition of immune cell infiltrates in precancerous colorectal lesions correlates with their morphology and/or size. Nonetheless, interesting insights can be gained by comparing our findings with those of the few studies that have investigated this phenomenon at a more general level. Among the stromal cell populations we quantified, CD4+ T helper cells were the most abundant in small adenomas (Fig 3), and their densities increased significantly with size (1.9- and 1.5-fold in large lesions from the polypoid and nonpolypoid groups, respectively). In colorectal cancer, progression from stage T1 to T4 is reportedly accompanied by a gradual decline in stromal counts of CD3+ cells, which include those that are CD4+ and/or CD8+ [20]. However, stromal CD4+ cell densities in invasive adenocarcinomas of the colon are significantly higher than those found in adenomas, and these cells are even less common in the normal mucosa [25]. These findings point to a progressive increase in the presence of CD4+ cells across the normal mucosa-adenoma-carcinoma sequence. This is fully consistent with our data showing higher CD4+ T-cell densities in larger and more dysplastic adenomas (Fig 3). More detailed sub-type level characterization of the CD4+ cell infiltrates is needed to define the biological and clinical significance of this trend: the T helper (Th) 1 and Th2 subsets have opposite effects on tumor growth, and Th1:Th2 ratios can change radically during tumor progression [18, 26].
Regulatory CD4+ T cells (Treg) can be reliably and specifically identified and quantified on the basis of FOXP3 expression. Tumor-infiltrating Treg cells dampen the anti-tumor immune response and promote tumor escape by producing immune-suppressive cytokines, adenosine, and prostaglandin E2 [40, 41]. This is consistent with reports of increasing Treg cell densities in colorectal tumors during progression through the precancerous and early cancerous stages [26, 28]. This trend was not reflected in our data: unlike CD4+ cell densities, Treg density did not increase significantly with lesion size. However, their abundance in the polypoid adenoma sections we examined was almost 3 times as high as that observed in the nonpolypoid lesions (Fig 3), and this feature might be expected to favor more rapid growth of polypoid tumors. It is important to recall, however, that tissue populations of Treg cells might include phenotypic and functional subsets, and their immunosuppressor activities might ultimately prove to be contextual. In invasive tumors, for example, heavy Treg cell infiltration is associated with a poor prognosis in ovarian cancer patients [42] and with better prognosis and improved overall survival in those with colorectal cancers [10]. Bindea et al. [20] found that colorectal cancer progression from T1 to T4 was accompanied by decreasing rather than increasing Treg cell densities. In the cancers they studied, the immune cells believed to mediate immunosuppression did not appear to exert any major tumor-promoting effects.
Cytotoxic (CD8+) T cells were also significantly more common in the polypoid adenomas we examined (fold change of 2.2 relative to nonpolypoid lesions) (Fig 3), and their density did not increase significantly with tumor size. In general, these cells were much less abundant than CD4+ T helper cells in adenomas, with CD8+/CD4+ ratios of 0.4 (small polypoid lesions), 0.3 (large polypoid lesions), and 0.23 (nonpolypoid lesions of all sizes). McLean et al. reported a CD8+/CD4+ ratio of 0.27 in adenomatous polyps in general (there was no differentiation based on morphology) and noted that the CD8+ cell density in these tumors resembled that observed in the adjacent normal mucosa [25]. Other investigators, however, maintain that the transition from normal colorectal mucosa to adenoma to cancer is associated with a progressive decline in stromal CD8+ T cell counts [10, 20, 28]. In light of our own data, we suspect that this latter trend might be more characteristic of nonpolypoid adenomas and that stable CD8+ cell densities across the normal mucosa-adenoma transition might be more typical of polypoid adenomas.
Stromal macrophages are key components of the tumor microenvironment that can influence tumor immunoediting (although their roles in the normal lamina propria of the gut are primarily phagocytic and bactericidal) [43]. In the stroma of our adenomas, macrophages (CD68+) were slightly more numerous than CD8+ or FOXP3+ T cells, but they were far less abundant than CD4+ T cells. Stromal macrophage densities were also significantly higher in polypoid lesions, although within the nonpolypoid group mild increases were observed with lesion size (Fig 3).
The functional heterogeneity and phenotypic plasticity of macrophage populations is even greater than that of other immune cell types. The two main polarization-based subtypes have more or less opposite functions in tumorigenesis: M1 macrophages are believed to exert anti-tumor effect by promoting the Th1 immune response; M2 macrophages favor the Th2 immune response, which facilitates tumor progression [44]. We used CD163 antibodies to quantitatively assess the latter tumor-promoting macrophage fraction in our colorectal adenomas. In both polypoid and nonpolypoid lesions, CD163+ cell densities were similar to those of CD68+ macrophages (Fig 3).
The predominance of M2 cells in the intratumoral macrophage population in these benign tumors was an unexpected finding, and it may not accurately reflect the characteristics of this early stage of tumorigenesis (i.e., presumably before the phase of tumor escape). CD163 is widely considered an M2-specific marker (reviewed in [15]), but Barros et al. [45] maintain that the macrophage polarization status can be more accurately classified on the basis of CD68 or CD163 expression plus transcription factor markers. Nevertheless, tumor infiltration by CD68+ macrophages tends to increase progressively as colorectal adenomas arise, grow, and become increasingly dysplastic ([23–25] and in our nonpolypoid lesions—Fig 3), and this trend seems to continue in the T1 and T2 stages of colorectal cancer [20]. However, immunohistochemistry studies by Edin et al. [19] indicate that colorectal cancer stage is inversely correlated with the densities of both M1 and M2 macrophages (identified by positivity for nitric oxide synthase 2 and CD163, respectively). Discordant findings have also emerged on the correlation between tumor-infiltrating macrophage densities and colorectal cancer prognosis or survival (reviewed in [15]).
MHC class I molecules are expressed by most nucleated cells. We used this marker only to obtain an overall count of stromal cells, a large proportion of which can reasonably be expected to be immune cells, including those not covered by other markers in our panel. Its nonspecificity for immune cells obviously precludes any speculations on the significance of our MHC-I+ cell counts in the process of immunoediting. Nonetheless, these counts provide complementary information on the picture that emerges based on expression patterns of the other 5 markers, i.e., that polypoid adenomas are characterized by more intense immune-cell stromal infiltration than their nonpolypoid counterparts. This difference might be related to the axial growth pattern of polypoid lesions, which protrude into the gut lumen. Compared with the lateral spread typical of nonpolypoid lesions, axial growth involves greater ramification and lengthening of the crypts. This type of growth is likely to require a more structured stromal support system, more replete with fibroblasts and vascular endothelial cells that would be reflected in higher MHC-I+ stromal cell counts.
The volume of a polypoid lesion is approximately twice that of a nonpolypoid with the same diameter, and the interstitial space available for immune / epithelial cell interaction is thus much larger. Early-stage polypoid colorectal cancers have more extensive microvascular networks than their nonpolypoid counterparts [46]. A similar difference might exist in precancerous lesions, and this facilitate the immune cells’ infiltration of the stroma of polypoid adenomas. The epithelial surface area might also play a role. Grivennikov et al. have suggested that the barrier function of adenomatous epithelium is compromised relative to that of the normal mucosa) [47]. If so, the larger surface area of a polypoid lesion (roughly twice that of a nonpolypoid adenoma with the same diameter) might result in a higher density of intralesional microbial antigens, which would trigger a more intense immune response. In our series and others, large nonpolypoid lesions (>10mm diameter) were more common in the proximal colon (Table 4), and tumor infiltration by immune cells might also be influenced by luminal factors peculiar to this colon segment. Serrated histology is also a common feature of nonpolypoid lesions, particularly those located in the proximal colon [48], but this factor had no effect on our findings since all but one of the lesions we studied were conventional adenomas (Table 4).
Table 4. Characteristics of the 40 colorectal adenomas included in the immunohistochemistry study.
| Patienta | Age | Sex | Colorectal segment involvedb | Maximum lesion diameter [mm] | Macroscopic appearancec | Dysplasiad |
|---|---|---|---|---|---|---|
| Small polypoid lesions | ||||||
| 1 | 37 | M | S | 16 | Ip | low |
| 2 | 74 | M | D | 15 | Ip | low |
| 3 | 74 | M | S | 16 | Ip | high |
| 4 | 58 | M | T | 15 | Ip | low |
| 5 | 63 | F | S | 15 | Ip | low |
| 6 | 60 | F | S | 15 | Is | low |
| 7 | 68 | M | S | 16 | Ip | low |
| 8 | 59 | M | S | 15 | Ip | low |
| 9 | 79 | F | S | 15 | Ip | low |
| 10 | 63 | F | S | 15 | Ip | high |
| Large polypoid lesions | ||||||
| 11 * | 68 | F | S | 30 | Ip | low |
| 12 * | 83 | M | S | 40 | Ip | low |
| 13 | 72 | F | S | 30 | Ip | high |
| 14 * | 64 | M | T | 50 | Ip | low |
| 15 | 57 | F | S | 35 | Ip | high |
| 16 | 84 | M | S | 35 | Is | high |
| 17 | 78 | F | S | 45 | Is | high |
| 18 | 88 | M | S | 30 | Is | high |
| 19 | 78 | F | R | 30 | Ip | high |
| Small nonpolypoid lesions | ||||||
| 20 | 58 | F | D | 18 | IIa | low |
| 21 | 52 | F | S | 15 | IIa | low |
| 22 | 83 | M | A | 15 | IIa | high |
| 23 | 62 | F | A | 15 | IIa | low |
| 24 | 66 | M | A | 18 | IIa | low |
| 25 | 63 | M | T | 18 | IIa | low |
| 26 | 56 | F | S | 15 | IIa | low |
| 27 | 62 | F | A | 15 | IIa | low |
| 28 | 73 | F | R | 15 | IIa | low |
| 29 | 69 | F | A | 15 | IIa | SSA |
| Large nonpolypoid lesions | ||||||
| 30 | 86 | M | D | 30 | IIa | low |
| 31 | 51 | F | C | 30 | IIa | low |
| 32 | 59 | M | A | 40 | IIa | high |
| 33 | 82 | F | D | 30 | IIa | high |
| 34 | 83 | F | A | 40 | IIa | high |
| 35 | 59 | F | D | 30 | IIa | high |
| 36 | 65 | F | D | 30 | IIa | low |
| 37 | 58 | M | D | 35 | IIa | high |
| 38 | 66 | F | R | 35 | IIa | high |
| 39 | 67 | F | T | 45 | IIa | low |
| 40 | 79 | M | A | 30 | IIa | high |
a * CD4 staining was not available these lesions
b Abbreviations: C, cecum; A, ascending colon; T, transverse; D, descending colon; S, sigmoid; R, rectum
c Classified according to the Paris Endoscopic Classification of Superficial Neoplastic Lesions [29]
d Highest degree of dysplasia in the lesion based on the WHO classification of tumors of the digestive system (Editorial and consensus conference in Lyon, France, November 6–9, 1999 [IARC]). SSA: sessile serrated adenoma.
Conclusions
An obvious limitation of our study is the restricted panel of immune cell markers we used and the fact that each was investigated individually (i.e. one antibody per histologic section). The complexity of the immune landscape in these lesions would undoubtedly be captured more efficiently by less reductionist methods (e.g., recently developed high-throughput techniques, such as multicolor immunohistochemistry and immunofluorescence, multiparameter flow cytometry, and transcriptomic and proteomic analyses).
The differences we have documented between the immune infiltrates of precancerous colorectal tumors with different morphologies may have implications for tumor immuno-chemoprevention. For example, the ability of aspirin and other nonsteroidal anti-inflammatory agents to reduce the recurrence of colorectal adenomas (reviewed in [5]) has been attributed by some to these drugs’ lowering of prostaglandin E2 levels, whose production in colorectal tumors is governed by interactions between epithelial and stromal cells [41, 49]. Therefore, significant differences in the immune cell infiltrates found in the stroma of polypoid and nonpolypoid adenomas might cause these lesions to respond differently to this type of intervention.
Materials and Methods
Colorectal tissue samples
The first series of human colorectal tissues analyzed in this study has been extensively described in a previous report [50], which includes an overview of the transcriptomic changes occurring during the course of tumor development. It consisted of 42 precancerous lesions, each with a sample of normal mucosa removed from the same colon segment at a distance of >2 cm from the tumor. The tissues were prospectively collected during diagnostic colonoscopy and their transcriptomes analyzed with the GeneChip Human Exon 1.0 ST array (Affymetrix, Santa Clara, CA, USA). Raw microarray data are available in the Gene Expression Omnibus repository (Series GSE21962). Transcript levels were expressed as log2 lesion and normal mucosa ratios. For our analyses, the 42 lesions were classified according to endoscopic morphology (polypoid [type Ip] vs. nonpolypoid [type IIa]) and size (small [11–20 mm] vs. large [>20 mm]) [29].
The second series of samples was used for immunohistochemistry studies. It included FFPE blocks of 40 precancerous colorectal lesions from the Cremona Hospital Pathology Department archives (Table 4). One of the nonpolypoid lesions displayed the histologic features of a sessile serrated adenoma/polyp; the other 39 were adenomas with some degree of dysplasia. As expected, most of the nonpolypoid lesions came from the proximal colon (see Introduction). The marginal normal mucosa of each FFPE block was not suitable for cell counting due to cauterization artifacts. The tumors in this archival series were deliberately selected to produce two distinct, non-overlapping size classes: small (diameters measuring from 15 to <20 mm) and large (diameters of ≥30 mm). Intermediate size (20–29 mm) lesions were excluded to increase the chance of identifying biological differences between early (small) and more advanced (large) adenomas. Therefore, our size classes in these experiments are not based on the 10-mm cutoff commonly used to distinguish small polyps from advanced lesions requiring closer endoscopic follow-up [30]. Indeed, lesions measuring < 10 mm have been excluded from all of the studies we have conducted thus far to ensure that our sampling procedure had no negative effects on the histologic diagnosis.
Colorectal tumors with defective DNA mismatch repair are usually characterized by more abundant lymphocyte infiltrates than their mismatch repair-proficient counterparts. None of the patients whose tissues were examined in this study had family histories indicative of a predisposition to this type of tumor. In addition, none had ever been diagnosed with sporadic mismatch repair defective tumors, which are associated with transcriptional silencing of MLH1. DNA from the lesions we studied was not analyzed for colon cancer gene mutations.
Recruitment and analyses of both sample series were approved by the Ethics Committees of the Hospital of Cremona (Italy). Each donor provided written informed consent to collection and analysis of data and publication of the findings.
Immunity-related transcriptomic (“immunome”) analysis (first series of 42 precancerous lesions)
Our analysis of transcriptomic data focused on the C7 immunologic signatures collection in the MSigDB, which is part of the Human Immunology Project Consortium (HIPC; http://www.immuneprofiling.org). GSEA was used to quantify immune-pathway upregulation or downregulation related to the morphology and size of the precancerous lesions. GSEA uses a variation of a Kolmogorov-Smirnov statistic to provide an enrichment score for each gene set. We use the signal-to-noise metric in the standard GSEA setting for measuring the correlation of a gene with the phenotype. The enrichment scores were then normalized to take into account the size of the gene sets resulting in a normalized enrichment score. This normalization was done using phenotypic permutations followed by standardization [39]. P-values and false discovery rates were computed using standard setting in the software. A p-value cut-off of 0.05 was used to define significant pathway enrichment.
Tissue staining for immunologic markers (2nd series of 40 precancerous lesions)
Shortly after removal, tissues were fixed in neutral buffered formalin (pH 7.0), dehydrated, and embedded in paraffin using standard histological techniques. Sections from FFPE blocks were stained with hematoxylin-eosin and periodic acid-Schiff stain for routine histologic analysis. Serial sections (5-micron) were processed for immunohistochemistry on Leica BondMax instruments using Refine HRP-kits (Leica DS9800), including all buffer solutions from Leica Microsystems Newcastle. Antibody characteristics and immunostaining procedures are summarized in Table 5. A Reichert microscope with a digital camera (JTV Polyvar 2) and Sony Trinitron monitor were used for quantitative analysis of immunostained cells. Labelled cells were counted in 10 microscopic fields (each measuring 140 x 110 microns, total amplitude: 15,400 square microns) at 400X magnification.
Table 5. Antibodies used in the immunohistochemistry study.
| Cell type | Cell Marker | Type | Antigen retrieval method * | Positive tissue control | Dilution | IHC detection protocol ** | Supplier | Code | Isotype / Clone |
|---|---|---|---|---|---|---|---|---|---|
| T helper cells | CD4 | Rabbit monoclonal | H2 20/95°C | Tonsil | 1:100 | rabbit HRP | Cell Marque Lifescreen | CMC10431021 | IgG / SP35 |
| cytotoxic T cells | CD8 | Rabbit monoclonal | H2(40) | Tonsil | 1:500 | rabbit HRP | Cell Marque Lifescreen | CMC1083100 | IgG / SP16 |
| T regulatory | FOXP3 | Rabbit monoclonal | H2(60) | Tonsil | 1:200 | rabbit HRP | Acris Antibodies | AM21067PU-M | IgG / SP97 |
| macrophages | CD68 | Mouse monoclonal | H2(30) | Tonsil | 1:200 | refine HRP | Novocastra Laboratories | NCL-L-CD68 | IgG2a_Kappa / 514H12 |
| macrophages | CD163 | Mouse monoclonal | H2(30) | Tonsil | 1:750 | refine HRP | Serotec | MCA1853 | IgG1 / EDHu-1 |
| nucleolated cells | MHC-I | Rabbit monoclonal | H1(30) | Tonsil | 1:500 | rabbit HRP | Epitomics | 2307–1 | IgG / EPR1394Y |
* Leica Bond Retrieval Buffer ER1 (30 min. at 100°C); Retrieval Buffer ER2 (for 30, 40 or 60 min at 100°C); Retrieval-Buffer ER2 for 20 min at 95°C.
** Rabbit HRP = Bond Polymer Refine HRP Kit from Leica (DS9800) without Postprimary Antibody (Rabbit anti-Mouse); Refine HRP: same Leica kit with Postprimary Antibody (Rabbit anti-Mouse)
MATLAB software (MathWorks, Natick, MA) was used for statistical analysis of the immunohistochemistry data. Non-parametric Wilcoxon Mann-Whitney tests were used to assess differences between mean values of immune cells in colorectal lesions according to their morphology, size, and degree of dysplasia. The significance level was set at p-value less than 0.05. MANOVA was used for testing the hypothesis that, given the sets of adenomatous lesions clustered in accordance to their morphology, size and degree of dysplasia, the means of the multiple response variable are or not distinct among the clusters of lesions, and the test is used to assess the statistical significance of the separation obtained among the lesions groups [51]. The null hypothesis H0 was that the two lesions groups came from the same population. MANOVA is accomplished by creating new variables (termed canonical) that maximize the differences among lesions of different morphology and/or size or degree of dysplasia. The first canonical variable is the linear combination of the original variables that best summarizes the differences among the lesion groups. The second canonical variable is the next best linear combination orthogonal to the first one, and so on. The grouped scatter plot of the first two canonical variables were used for displaying group structure in our data. Another graphical output of this analysis was the dendogram plot of the group mean clusters following MANOVA, where the clusters were computed by applying the single linkage method to the matrix of Mahalanobis distances between group means.
Supporting Information
A total of 429 pathways (graphically represented in Fig 1) displayed significant up- or downregulation (p-value < 0.05) in GSEA of one or more of the five group comparisons: 1) polypoid vs. nonpolypoid (all sizes); 2) polypoid vs. nonpolypoid (small lesions only); 3) polypoid vs. nonpolypoid (large lesions only); 4) small vs. large (polypoid lesions only); 5) small vs. large (nonpolypoid lesions only). The column degree shows the number of group comparisons in which significant differential regulation of the pathway was found.
(XLSX)
Acknowledgments
We are very grateful to M. van den Broek for helpful suggestions and comments, A. Argentieri, R. Colella and S. Behnke for technical assistance, M.E. Kent for translation and editing, and P. Soria for graphical work.
Abbreviations
- GSEA
Gene Set Enrichment Analysis
- MSigDB
Molecular Signature Database
- FFPE
formalin-fixed paraffin-embedded
- MANOVA
One-way Multivariate Analysis of Variance
- Treg
T regulatory cells
Data Availability
Raw microarray data are available in the Gene Expression Omnibus repository (Series GSE21962). URL: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE21962.
Funding Statement
The research was partially supported by a grant of the Swiss National Science Foundation to GM (no. 31003A_141225), of Regione Puglia to NA (PO FESR 2007-2013 Progetto BISIMANE Cod. n. 44, Progetto FIRB RBAP11B2SX, Progetto di Ricerca Finalizzata 2009 RF/2009-1471624). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Schreiber RD, Old LJ, Smyth MJ. Cancer immunoediting: integration immunity’s roles in cancer suppression and promotion. Science 2011, 331:1565 10.1126/science.1203486 [DOI] [PubMed] [Google Scholar]
- 2.Stryker SJ, Wolff BG, Culp CE, Libbe SD, Ilstrup DM, MacCarty RL. Natural history of untreated colonic polyps. Gastroenterology 1987, 93(5):1009–1013 [DOI] [PubMed] [Google Scholar]
- 3.Brenner H, Hoffmeister M, Stegmaier C, Brenner G, Altenhofen L, Haug U. Risk of progression of advanced adenomas to colorectal cancer by age and sex: estimates based on 840 149 screening colonoscopies. Gut. 2007, 56:1585–1589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cattaneo E, Baudis M, Buffoli F, Bianco MA, Zorzi F, Marra G. Pathways and crossroads to colorectal cancer. Pre-Invasive Disease: Pathogenesis and Clinical Management 2011, 369–394 [Google Scholar]
- 5.Thun MJ, Jacobs EJ, Patrono C. The role of aspirin in cancer prevention. Nature Review Clinical Oncology 2012, 9:259–267 [DOI] [PubMed] [Google Scholar]
- 6.Chia WK, Ali R, Toh HC. Aspirin as adjuvant therapy for colorectal cancer—reinterpreting paradigms. Nature Review Clinical Oncology 2012, 9:561–570 [DOI] [PubMed] [Google Scholar]
- 7.Naito Y, Saito K, Shiiba K, Ohuchi A, Saigenji K, Nagura H et al. CD8+ T Cells Infiltrated within Cancer Cell Nests as a Prognostic Factor in Human Colorectal Cancer. Cancer Research 1998, 58:3491 [PubMed] [Google Scholar]
- 8.Menon AG, Janssen-van Rhijn CM, Morreau H, Putter H, Tollenaar RA, van de Velde CJ, et al. Immune system and prognosis in colorectal cancer: a detailed immunohistochemical analysis. Laboratory Investigation 2004, 84:493–501 [DOI] [PubMed] [Google Scholar]
- 9.Galon J, Costes A, Sanchez-Cabo F, Kirilovsky A, Mlecnic B, Lagorge-Pàges C, et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science 2006, 313:1960–1964 [DOI] [PubMed] [Google Scholar]
- 10.Salama P, Phillips M, Grieu F, Morris M, Zeps N, Joseph D, et al. Tumor- infiltrating FOXP3+ T regulatory cells show strong prognostic significance in colorectal cancer. J Clin Oncol 2009, 27:186–192 10.1200/JCO.2008.18.7229 [DOI] [PubMed] [Google Scholar]
- 11.Laghi L, Bianchi P, Miranda E, Balladore E, Pacetti V, et al. CD3+ cells at the invasive margin of deeply invading (pT3-T4) colorectal cancer and risk of post-surgical metastasis: a longitudinal study. The lancet oncology 2009, 10(9):877–84 10.1016/S1470-2045(09)70186-X [DOI] [PubMed] [Google Scholar]
- 12.Pages F, Kirilovsky A, Mlecnik B, Asslaber M, Tosolini M, Bindea G, et al. In Situ Cytotoxic and Memory T Cells Predict Outcome in Patients With Early-Stage Colorectal Cancer. Journal of Clinical Oncology 2009, 27:5944–5951 10.1200/JCO.2008.19.6147 [DOI] [PubMed] [Google Scholar]
- 13.Simpson JAD, Al-Attar A, Watson NFS, Scholefield JH, Ilyas M, Durrant LG. Intratumoral T cell infiltration, MHC class I and STAT1 as biomarkers of good prognosis in colorectal cancer. Gut 2010, 59:926–33 10.1136/gut.2009.194472 [DOI] [PubMed] [Google Scholar]
- 14.Terzić J, Grivennikov SI, Karin E, Karin M. Inflammation and colon cancer. Gastroenterology 2010, 138(6):2101–2114 10.1053/j.gastro.2010.01.058 [DOI] [PubMed] [Google Scholar]
- 15.Heusinkveld M, Van Der Burg SH. Identification and manipulation of tumor associated macrophages in human cancers. Journal of Translational Medicine 2011, 9:216 10.1186/1479-5876-9-216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ladoire S, Martin F, Ghiringhelli F. Prognostic role of FOXP3+ regulatory T cells infiltrating human carcinomas: the paradox of colorectal cancer. Cancer Immunol Immunother 2011, 60:909–918 10.1007/s00262-011-1046-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mlecnik B, Tosolini M, Kirilovsky A, Berger A, Bindea G, Meatchi T, et al. Histopathologic-Based Prognostic Factors of Colorectal Cancers Are Associated With the State of the Local Immune Reaction. Journal of Clinical Oncology 2011, 29:610 10.1200/JCO.2010.30.5425 [DOI] [PubMed] [Google Scholar]
- 18.Tosolini M, Kirilovsky A, Mlecnik B, Fredericksen T, Mauger S, Bindea G, et al. Clinical impact of different classes of infiltrating T cytotoxic and helper cells (Th1, Th2, Treg, Th17) in patients with colorectal cancer. Cancer Res 2011, 71:1263–71 10.1158/0008-5472.CAN-10-2907 [DOI] [PubMed] [Google Scholar]
- 19.Edin S, Wikberg ML, Dahlin AM, Rutegård J, Öberg Å, Oldenborg PA, et al. The Distribution of Macrophages with a M1 or M2 Phenotype in Relation to Prognosis and the Molecular Characteristics of Colorectal Cancer. PLoS ONE 2012, 7(10): e47045 10.1371/journal.pone.0047045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bindea G, Mlecnik B, Tosolini M, Kirilovsky A, Waldner M, Obenauf A, et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity 2013, 39:782–795 10.1016/j.immuni.2013.10.003 [DOI] [PubMed] [Google Scholar]
- 21.Grizzi F, Bianchi P, Malesci A, Laghi L. Prognostic value of innate and adaptive immunity in colorectal cancer World J Gastroenterol 2013, 19:174–184 10.3748/wjg.v19.i2.174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yuan A, Steigen SE, Goll R, Vonen B, Husbekk A, Cui G, et al. Dendritic cell infiltration pattern along the colorectal adenoma- carcinoma sequence. APMIS 2008, 116: 445–56 [PubMed] [Google Scholar]
- 23.Cui G, Yuan A, Vonen B, Florholmen J. Progressive cellular response in the lamina propria of the colorectal adenoma-carcinoma sequence. Histopathology 2009, 54:550 10.1111/j.1365-2559.2009.03273.x [DOI] [PubMed] [Google Scholar]
- 24.Cammarota R, Bertolini V, Pennesi G, Bucci EO, Gottardi O, Garlanda C, et al. The tumor microenvironment of colorectal cancer: stromal TLR-4 expression as a potential prognostic marker. Journal of Translational Medicine 2010, 8:112 10.1186/1479-5876-8-112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McLean MH, Murray GI, Stewart KN, Norrie G, Mayer C, Hold GL, et al. The inflammatory microenvironment in colorectal neoplasia. PLoS ONE 2011, 6(1): e15366 10.1371/journal.pone.0015366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cui G, Shi Y, Cui J, Tang T, Florholmen J. Immune microenvironmental shift along human colorectal adenoma–carcinoma sequence: is it relevant to tumor development, biomarkers and biotherapeutic targets? Scandinavian Journal of Gastroenterol 2012, 47:367. [DOI] [PubMed] [Google Scholar]
- 27.Mariani F, Sena P, Pedroni M, Benatti P, Manni P, Di Gregorio C, et al. Th inducing POZ- Kruppel Factor (ThPOK) is a key regulator of the immune response since the early steps of colorectal carcinogenesis. PLos ONE 2013, 8(1): e54488 10.1371/journal.pone.0054488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jang TJ. Progressive Increase of Regulatory T Cells and Decrease of CD8+ T Cells and CD8+ T Cells/Regulatory T Cells Ratio during Colorectal Cancer Development. The Korean Journal of Pathology 2013, 47:443 10.4132/KoreanJPathol.2013.47.5.443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Endoscopic Classification Review Group. Update on the Paris classification of superficial neoplastic lesions in the digestive tract. Endoscopy 2005, 37:570–578 [DOI] [PubMed] [Google Scholar]
- 30.Lieberman DA, Rex DK, Winawer SJ, Giardiello FM, Johnson DA, Levin TR, et al. Guidelines for colonoscopy surveillance after screening and polypectomy: a consensus update by the US Multi-Society Task Force on Colorectal Cancer. Gastroenterology 2012, 143(3):844–57 10.1053/j.gastro.2012.06.001 [DOI] [PubMed] [Google Scholar]
- 31.Bettington M, Walker N, Clouston A, Brown I, Legget B, Whitehall V: The serrated pathway to colorectal carcinoma: current concepts and challenges. Histopathology 2013, 62:367–386 10.1111/his.12055 [DOI] [PubMed] [Google Scholar]
- 32.O'Brien MJ, Winawer SJ, Zauber AG, Bushey MT, Sternberg SS, Gottlieb LS, et al. Flat adenomas in the National Polyp Study: is there increased risk for high-grade dysplasia initially or during surveillance? Clinical Gastroenterology & Hepatology 2004, 2(10):905–11 [DOI] [PubMed] [Google Scholar]
- 33.Kudo S, Lambert R, Allen JI, Fujii H, Fujii T, Kashida H, et al. Nonpolypoid neoplastic lesions of the colorectal mucosa. Gastrointest Endosc 2008, 68(4Suppl):S3–47 [DOI] [PubMed] [Google Scholar]
- 34.Soetikno RM, Kaltenbach T, Rouse RV, Park W, Maheshwari A, Sato T, et al. Prevalence of nonpolypoid (flat and depressed) colorectal neoplasms in asymptomatic and symptomatic adults. JAMA 2008; 299:1027–1035 10.1001/jama.299.9.1027 [DOI] [PubMed] [Google Scholar]
- 35.Bianco MA, Cipolletta L, Rotondano G, Buffoli F, Gizzi G, Tessari F, et al. Prevalence of nonpolypoid colorectal neoplasia: an Italian multicenter observational study. Endoscopy 2010, 42(4):279–85 10.1055/s-0029-1244020 [DOI] [PubMed] [Google Scholar]
- 36.Park DH, Kim HS, Kim WH, Kim TI, Kim YH, Park DI, et al. Clinicopathologic characteristics and malignant potential of colorectal flat neoplasia compared with that of polypoid neoplasia. Dis Colon Rectum 2008, 51:43–9 [DOI] [PubMed] [Google Scholar]
- 37.Voorham QJM, Rondagh EJ, Knol DL, van Engeland M, Carvalho B, Meijer GA, et al. Tracking the Molecular Features of Nonpolypoid Colorectal Neoplasms: A Systematic Review and Meta-Analysis. American Journal of Gastroenterology 2013, 108:1042 10.1038/ajg.2013.126 [DOI] [PubMed] [Google Scholar]
- 38.Cattaneo E, Laczko E, Buffoli F, Zorzi F, Bianco MA, Monigatti M, et al. Preinvasive colorectal lesion transcriptomes correlate with endoscopic morphology (polypoid vs. nonpolypoid). EMBO Mol Med 2011, 3(6):334–47 10.1002/emmm.201100141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Subramanian A, Tamayo P, Mootha V, Mukherjee S, Ebert B, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles Proc Natl Acad Sci 2005, 102:15545–15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gallimore A, Godkin A. Regulatory T cells and tumour immunity–observations in mice and men. Immunology 2008, 123:157–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Whiteside TL. Regulatory T cell subsets in human cancer: are they regulating for or against tumor progression? Cancer Immunol Immunother 2014, 63: 67–72 10.1007/s00262-013-1490-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Curiel TJ, Coukos G, Zou L, Alvarez X, Cheng P, Mottram P, et al. Specific recruitment of regulatory T cells in ovarian carcinoma fosters immune privilege and predicts reduced survival. Nat Med 2004, 10:942–949 [DOI] [PubMed] [Google Scholar]
- 43.Schenk M, Mueller C. The mucosal immune system at the gastrointestinal barrier Best Practice & Research Clinical Gastroenterology 2008. 22(3): 391–409 10.1016/j.bpg.2007.11.002 [DOI] [PubMed] [Google Scholar]
- 44.Mantovani A, Sica A. Macrophages, innate immunity and cancer: balance, tolerance, and diversity. Current opinion in immunology 2010, 22:231–237 10.1016/j.coi.2010.01.009 [DOI] [PubMed] [Google Scholar]
- 45.Barros MHM, Hauck F, Dreyer JH, Kempkes B, Niedobitek G. Macrophage Polarisation: an Immunohistochemical Approach for Identifying M1 and M2 Macrophages. PLoS ONE 2013, 8(11): e80908 10.1371/journal.pone.0080908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Okada K, Satoh T, Fujimoto K, Tokunaga O. Interaction between morphology and angiogenesis in human early colorectal cancers. Pathology International 2004, 54: 490–497 [DOI] [PubMed] [Google Scholar]
- 47.Grivennikov SI, Wang K, Mucida D, Stewart CA, Schnabl B, Jauch D, et al. Adenoma-linked barrier defects and microbial products drive IL-23/IL-17-mediated tumor growth. Nature 2012, 491:254–258 10.1038/nature11465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rex DK, Ahnen DJ, Baron JA, Batts KP, Burke CA, Burt RW, et al. Serrated lesions of the colorectum: review and recommendations from an expert panel. Am J Gastroenterol 2012, 107:1315–1329 10.1038/ajg.2012.161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Greenhough A, Smartt HJM, Moore AE, Roberts HR, Williams AC, Paraskeva C, et al. The COX-2/PGE2 pathway: key roles in the hallmarks of cancer and adaptation to the tumour microenvironment. Carcinogenesis 2009, 30(3):377–386 10.1093/carcin/bgp014 [DOI] [PubMed] [Google Scholar]
- 50.Maglietta R, Liuzzi VC, Cattaneo E, Laczko E, Piepoli A, Panza A, et al. Molecular pathways undergoing dramatic transcriptomic changes during tumor development in the human colon BMC Cancer 2012, 12:608 10.1186/1471-2407-12-608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Krzanowski WJ. Principles of Multivariate Analysis: A User's Perspective New York: Oxford University Press; 1988 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A total of 429 pathways (graphically represented in Fig 1) displayed significant up- or downregulation (p-value < 0.05) in GSEA of one or more of the five group comparisons: 1) polypoid vs. nonpolypoid (all sizes); 2) polypoid vs. nonpolypoid (small lesions only); 3) polypoid vs. nonpolypoid (large lesions only); 4) small vs. large (polypoid lesions only); 5) small vs. large (nonpolypoid lesions only). The column degree shows the number of group comparisons in which significant differential regulation of the pathway was found.
(XLSX)
Data Availability Statement
Raw microarray data are available in the Gene Expression Omnibus repository (Series GSE21962). URL: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE21962.