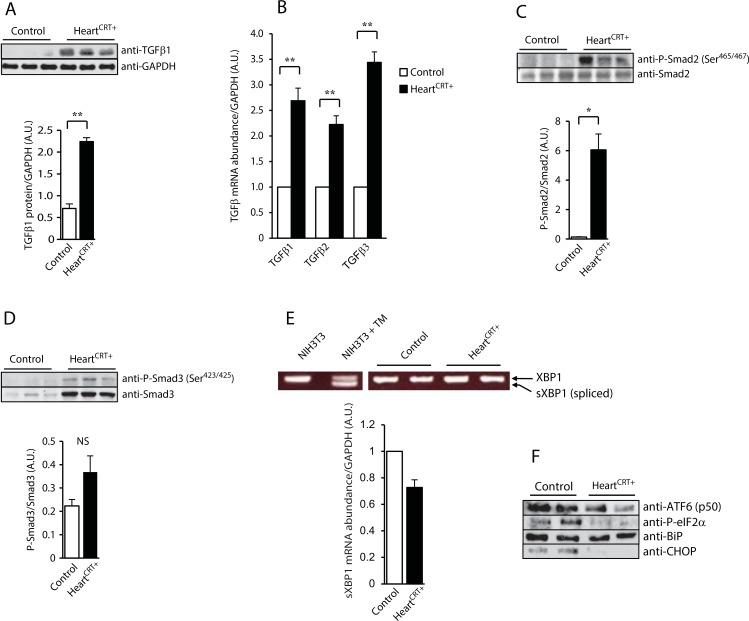
Fig 2. TGFβ1 and unfolded protein response (UPR) in calreticulin expressing hearts.
(A) Immnoblot analysis and quantification of TGF-β1 protein in control and HeartCRT+ hearts. Antibodies to glyceraldehyde 3-phosphate dehydrogenase (GAPDH) were used as a loading control. **p<0.01. Data are representative of 6 biological replicates. (B) Real-time Q-PCR analysis for TGF-β1, β2 and β3 transcripts. **p<0.01. Data are representative of 6 biological replicates. (C) Immunoblot analysis of Smad2 and phospho-Smad2 (P-Smad2) at Ser465 and Ser467 antibodies from control and HeartCRT+ hearts. Quantitative analysis of P-Smad2/Smad2 expression ratio shown in. Anti-GAPDH antibodies were used as a loading control. * p<0.05. Data are representative of 6 biological replicates. (D) Immnuoblot analysis of Smad3 and phospho-Smad3 (P-Smad3) at Ser423 and Ser425 antibodies from control and HeartCRT+ hearts. Quantitative analysis of P-Smad3/Smad3 ratio. Anti-GAPDH antibodies were used as a loading control. NS, not significant. Data are representative of 6 biological replicates. (E) Real-time Q-PCR analysis of spliced XBP1 (sXBP1) mRNA abundance. The abundance of the sXBP1 transcripts in HeartCRT+ was slightly decreased (0.73±0.06 vs. control). **p<0.01. Data are representative of 6 biological replicates. NIH3T3 cells treated with tunicamycin (TM), an activator of unfolded protein response, were used as a positive control for the XBP1 splicing analysis. (F) Immnunoblot analyses of UPR markers were carried out with specific anti-ATF6 p50 fragment, anti-P-elF2α, anti-BiP and anti-CHOP antibodies. Anti-GAPDH antibodies were used as a loading control. Data are representative of 6 biological replicates.