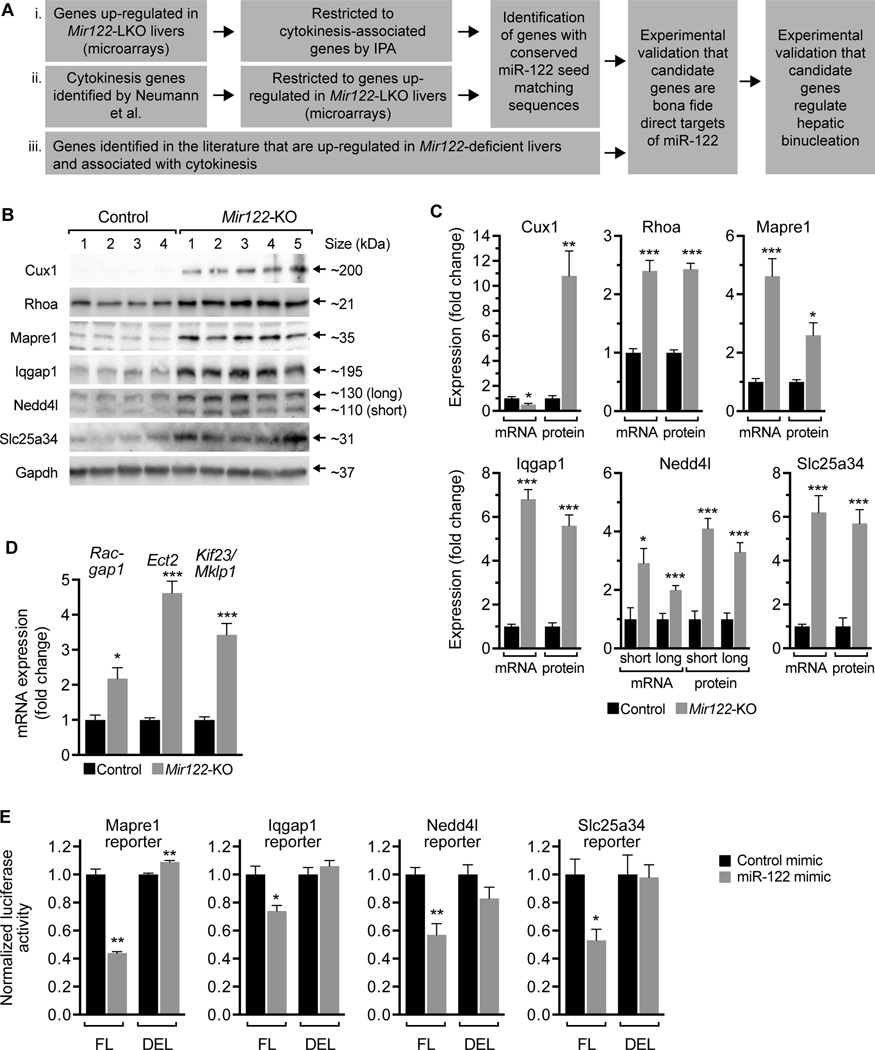
Fig. 5. miR-122 regulates expression of cytokinesis-related targets.
(A) Strategy for identifying miR-122 target genes that regulate cytokinesis. (B,C) Cux1, Rhoa, Mapre1, Iqgap1, Nedd4l and Slc25a34 were up-regulated in 28d-old Mir122-KO livers compared to the controls at the protein and/or mRNA levels (n = 4–5). Western blots are shown for 4 control and 5 Mir122-KO livers; Gapdh was used as a loading control (B). Graphs show relative mRNA and protein expression levels normalized to Gapdh (C). (D) Centralspindlin complex members Racgap1, Ect2 and Kif23/Mklp1 were also up-regulated in 28d-old Mir122-KO livers (n = 5). The graph shows mRNA expression normalized to Gapdh. (E) NIH 3T3 cells were transfected with Mapre1, Iqgap1, Nedd4l or Slc25a34 reporter plasmids containing predicted miR-122 3’UTR seed sequences (full length, FL) or with the seed sequences deleted (DEL). Reporter constructs are detailed in Supporting Fig. 8. Cells were co-transfected with control or miR-122 mimic (25 nM) and luciferase activity measured after 48h. For each set of reporters, miR-122 expression reduced reporter activity 26–47% in the presence of FL reporters but not DEL reporters. Graphs show normalized luciferase activity (Renilla luciferase activity controlled by FL or DEL normalized to firefly luciferase activity expressed by the same plasmid) (n = 3 replicates from 2–3 independent experiments). * P < 0.03, ** P < 0.008, *** P < 0.0008. Graphs show mean ± SEM.