Figure 1. Proteins, isoforms, proteoforms, and the naming convention of each.
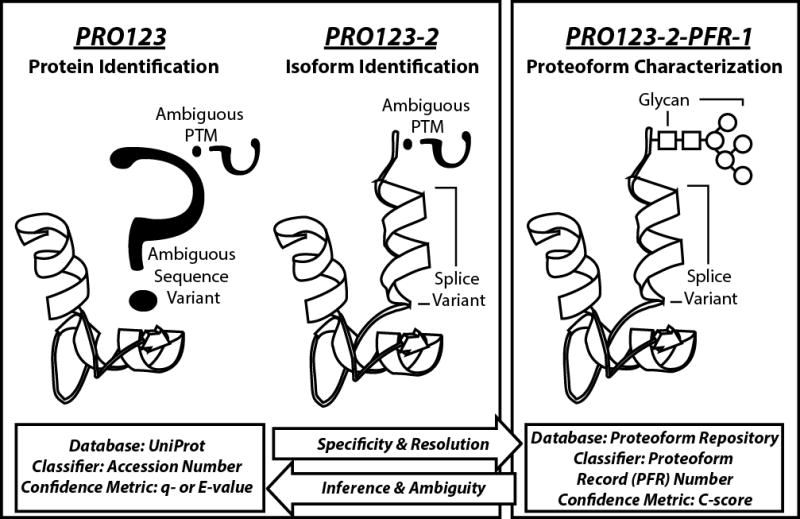
Protein molecules can be discussed using three levels of increasing specificity: protein identification (far left), isoform identification (middle), and proteoform characterization (far right). UniProtKB entries are based on accession numbers, which is gene-specific but does not distinguish between alternative transcripts or molecular variants resulting from post-translational modification (PTM). UniProt handles the alternative transcripts by introducing isoform information into the classification, but this is still unable to distinguish different forms of proteins carrying specific PTMs. The proteoform record (PFR) number carries maximal molecular specificity by referring to an intact protein molecule’s chemical formula, including type and location of PTMs. In top down proteomics, confidence metrics for protein identification by database retrieval are q- or E-values (bottom left); for proteoform characterization the metric is the C-score (bottom right). “PRO123” is a hypothetical protein; “PRO123-2” specifies a non-canonical isoform of a UniProt entry; “PFR-1” is a specific proteoform, which contains a glycan modification, and maps to the hypothetical PRO123 accession in UniProtKB.
