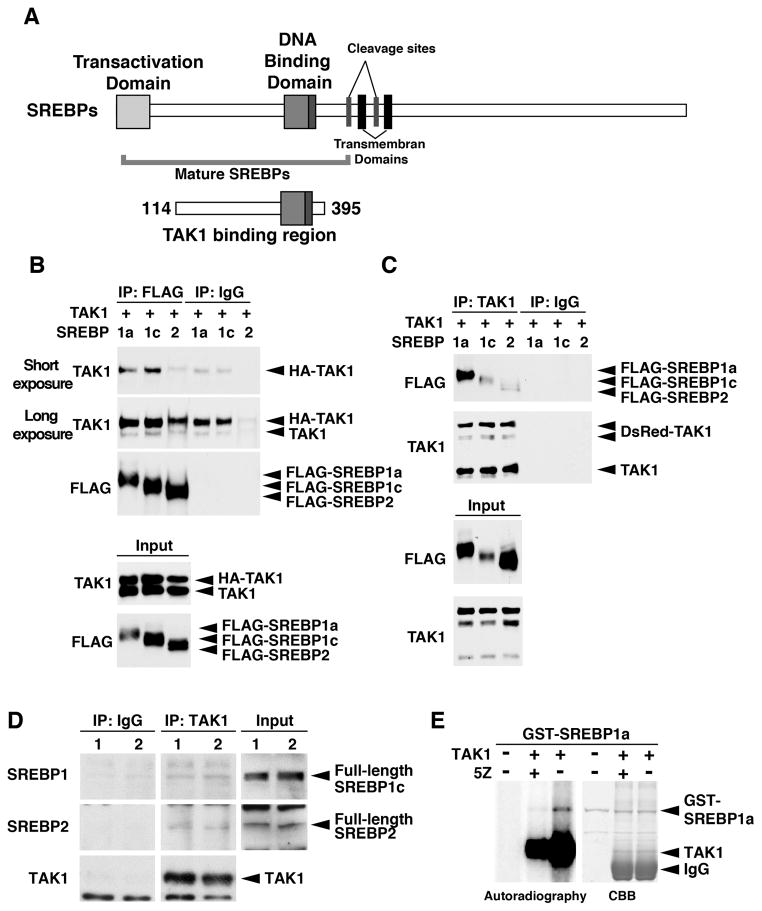
Figure 2. TAK1 interacts with SREBP.
(A) Structure of SREBPs. The region of TAK1 binding domain isolated from the yeast two-hybrid screening is denoted by encompassing amino acid residues of SREBP2. (B, C) HEK293 cells were transfected with expression vectors for HA- (B) or DsRed- (C) tagged TAK1 and FLAG-tagged mature forms of SREBPs (N-terminal region), and proteins from cell lysates were immuneprecipitated by anti-FALG (B) or anti-TAK1 (C) as well as non-immunized control IgG. Immunoprecipitates were analyzed by immunoblotting. The amounts of input proteins are also shown in bottom two panels. Shorter (B, top panel) and longer (B, second panel) exposures are shown to visualize less efficiently co-precipitated HA-TAK1 (3rd lane) and endogenous TAK1 (D) Cytoplasmic fractions including cytoplasmic organelle fractions from the mouse liver were immunoprecipitated by anti-TAK1 or control IgG. Immunoprecipitates were analyzed by immunoblotting. Each lane (lane 1 and 2) represents a sample from an individual mouse. (E) HEK293 cells were transfected with expression vectors for HA-tagged catalytic domain of TAK1 (TAK1ΔC) together with TAB1. HA-TAK1ΔC was purified from protein extracts by immunoprecipitation, and incubated with bacterially purified GST-tagged mature SREBP1a in the presence (+) or absence (−) of 5Z-7oxozeaenol (5Z) (500 nM). Phosphorylation of the proteins was detected by autoradiography. The protein amounts are shown by coomassie brilliant blue (CBB) staining.