Figure 5.
The DCBBIG1 Domain Dimer
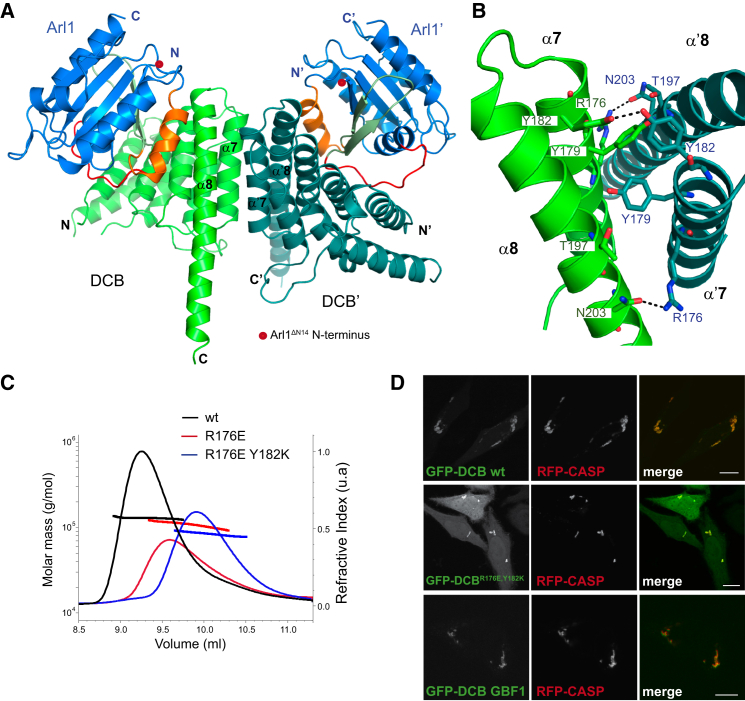
(A) Ribbon diagrams of two adjacent asymmetric units of the Arl1GTP/DCBΔ51–71 complex. The two DCB domains (green and turquoise) interact through helices α7 and α8. Each DCB domain binds to a molecule of Arl1 (blue, with switch 1 in red, interswitch in pale green, and switch 2 in orange) such that the two Arl1 N termini (red dots) are the same side of the complex.
(B) DCBΔ51–71 dimer interface. Residues involved in dimerization are shown as sticks and contacts based on hydrogen bonds with a distance below 3 Å (dashed lines).
(C) Determination of native molecular weight of MBP-tagged DCBBIG1 proteins by SEC-MALS. The expected size for a MBP-DCBBIG1 monomer is 69 kDa, while the average mass for the wild-type protein is 129 kDa, R176E mutant is 110 kDa, R176E/Y182K is 83 kDa.
(D) Confocal micrographs of live HeLa cells expressing the N-terminal GFP-tagged DCB domains from BIG1 or GBF1 as indicated. An RFP-tagged form of the golgin CASP was used as a Golgi marker (Gillingham et al., 2002). The R176E/Y182K mutant is expressed at slightly higher levels than the wild-type protein for reasons that are unclear. It is still targeted to the Golgi, and at comparable expression levels, the mutant appeared indistinguishable from wild-type, but we show representative cells where the expression levels, and hence cytoplasmic staining, are higher. Scale bars, 5 μm.