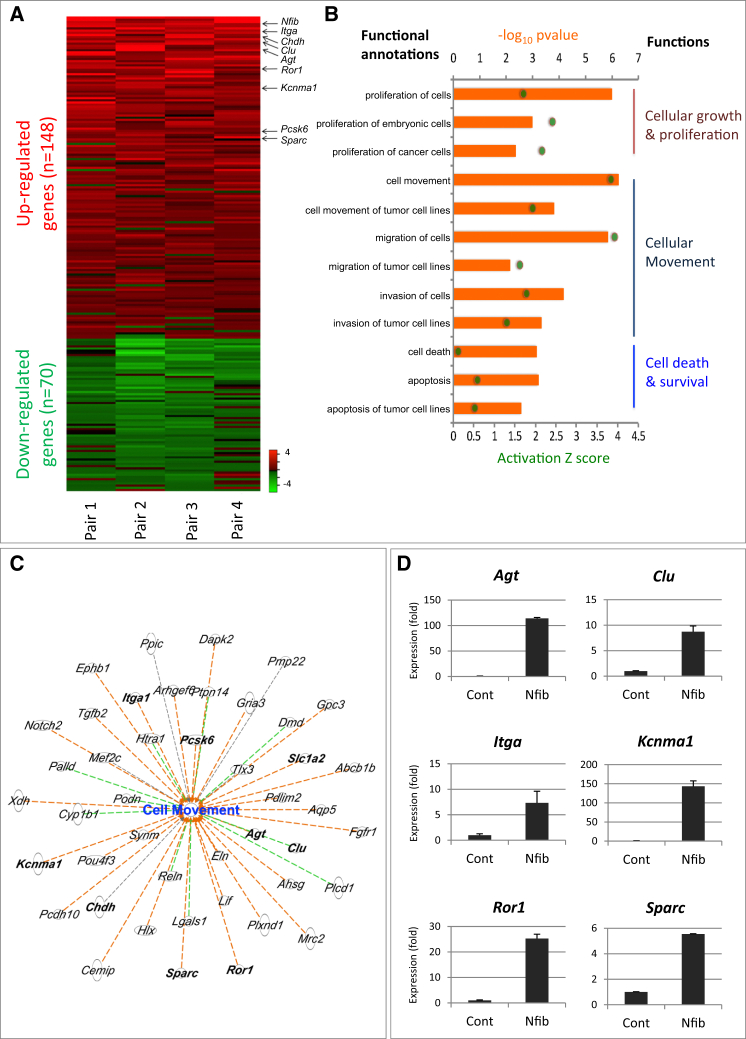
Figure 4.
Key Changes in Cancer Progression Following NFIB Overexpression In Vitro
(A) Analysis of the gene expression changes following Nfib overexpression in four mouse SCLC primary cell lines (heatmap). DEGs were selected if expressed in at least three out of four samples. Values are represented as log2 fold change. Red, upregulated (average log2 fold change > 0.5); green, downregulated genes (average log2 fold change < −0.5).
(B) Ingenuity pathway analysis (IPA) of differentially expressed genes based on Z score and p value. The biological functions that are expected to be increased according to the gene expression changes in our dataset were identified using the IPA regulation Z score algorithm (green dot). The p value was calculated with the Fischer’s exact test (p value ≤ 0.05).
(C) Genes involved in cell movement. Orange dashed line, predicting activation of cell movement; green dashed line, unknown directionality; black dashed line, effect not predicted.
(D) Validation of several genes in (C) by qPCR. Error bars represent mean ± SD.