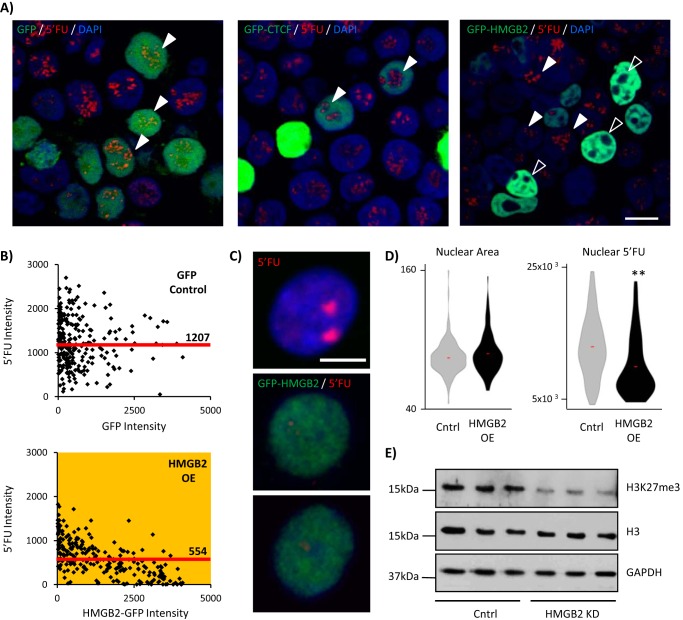
FIGURE 4.
hmgb2 overexpression represses transcription. A, control, hmgb2-gfp-overexpressing or ctcf-gfp-overexpressing 293T cells were treated with 5′FU to label newly transcribed RNA. hmgb2-overexpressing cells (green cells) exhibited a loss of nucleolar transcription (5′FU in red) that was seen with neither the ctcf overexpression nor gfp overexpression alone. Bar, 10 μm. Image representative of n >100; experiment was repeated four times. B, there was a significant (yellow indicates p value <0.001 (Mann-Whitney)) inverse relationship between the intensity of Hmgb2-GFP in the nuclei, and the intensity of the 5′FU signal, which was not seen in the gfp-only treatment (n = top 250 cells with most Gfp intensity; significance was also true when compared with all n = 329 cells measured; median 5′FU intensity is indicated by labeled red line; one representative experiment of three). C, we repeated these analyses in NRVMs after 30 min of 5′FU labeling (red) in control (top panel) and hmgb2-gfp-overexpressing (middle and bottom panels) cells (bar, 5 μm), confirming that hmgb2 overexpression decreases total 5′FU nuclear signal without affecting nuclear size (** indicates p value <0.0001; one representative experiment of three; D). E, Western blotting confirmed a decrease in tri-methylation of lysine 27, a mark of facultative heterochromatin, supporting a role for large scale changes in Hmgb2 concentration to modulate global transcriptional levels (n = 3, blots quantified in ImageJ; H3K27me3 normalized to H3 gave mean signal of 1.03 for Lipofectamine control (S.D. 0.16) versus 0.12 for hmgb2 knockdown (S.D. 0.04), p value = 0.0006 (two-tailed t test; one representative experiment of three).