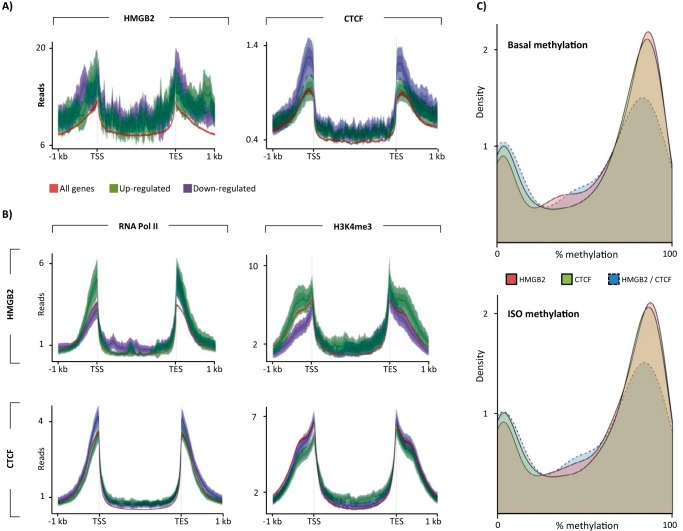
FIGURE 6.
Opposing effects of local chromatin environment on Hmgb2 versus Ctcf-regulated genes. A, Hmgb2 reads or Ctcf reads were plotted across genes up-regulated or down-regulated by hmgb2 knockdown or ctcf knock-out (left and right panels, respectively) or across all genes in the genome, revealing that the distribution of Hmgb2/Ctcf is not the major determinant of whether a gene's expression will change with depletion of these proteins (Hmgb2- and Ctcf-regulated genes use expression data averaged from three experiments). B, distribution of ChIP-seq (ENCODE, basal adult mouse heart) reads across hmgb2-regulated genes suggests genes that are up-regulated by knockdown are enriched in the activating marks H3K4me3 and RNA pol II in the basal state as compared with genes that are down-regulated by knockdown (top panel). Strikingly, the opposite is true for genes that are regulated by ctcf (bottom panel; ENCODE data represents one biological replicate). C, we next examined DNA methylation from the mouse heart. Plotted is the bimodal distribution of average DNA methylation across Hmgb2-only and Ctcf-only peaks in intergenic regions. Shared peaks are less likely to have high methylation but rather show low or intermediate level methylation. This is true using DNA methylation data from basal adult mice and mice treated with isoproterenol for 3 weeks (DNA methylation is average of six mice (54)).