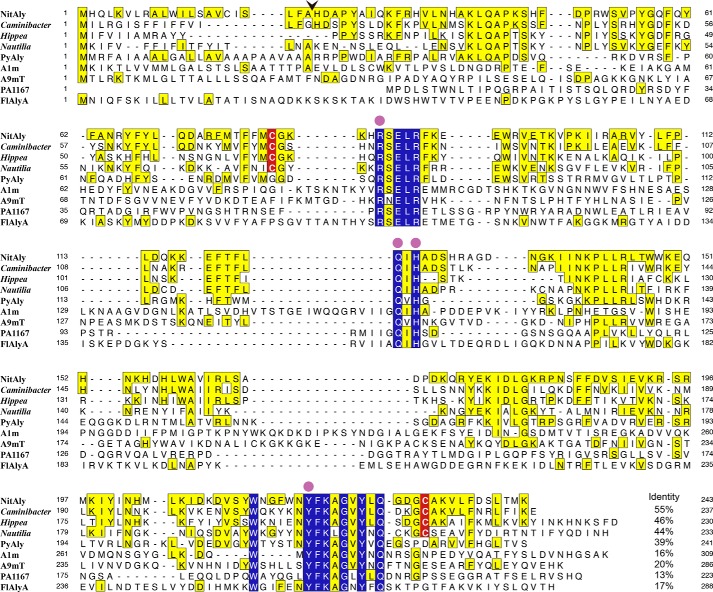
FIGURE 1.
Comparison among the amino acid sequences of NitAly, its homologous proteins, and several characterized family PL-7 alginate lyases. NitAly, Nitratiruptor sp. SB155-2 hypothetical protein NIS_0185 (GenBankTM accession no. WP_012081562); Caminibacter, C. mediatlanticus hypothetical protein (GenBankTM accession no. WP_007473671); Hippea, H. jasoniae hypothetical protein (GenBankTM accession no. WP_051904545); Nautilia, N. profundicola predicted alginate lyase (GenBankTM accession no. WP_015901805); PyAly, P. yezoensis alginate lyase PyAly (GenBankTM accession no. BAI66416) (39); PA1167, P. aeruginosa PAO1 alginate lyase PA1167 (GenBankTM accession no. AAG04556) (15); A1m, Agarivorans sp. JAM-A1m alginate lyase A1m (GenBankTM accession no. AB426616) (31); A9mT, Vibrio sp. alginate lyase A9mT (GenBankTM accession no. AB473598) (25); FlAlyA, Flavobacterium sp. UMI-01 alginate lyase FlAlyA (18). Residues invariant among all listed proteins are indicated with blue boxes. Residues identical with those of NitAly are indicated with yellow boxes. Cys-80 and Cys-232 in NitAly and their corresponding conserved Cys residues in other proteins are highlighted with red. An arrowhead shows the cleavage site of the signal peptide in NitAly predicted by the SignalP 3.0 software program. Catalytic residues proposed in PL-7 alginate lyases (15, 21, 65) are shown by solid circles. The level of identity of each other protein with NitAly is shown at the end of each sequence, respectively.