FIGURE 8.
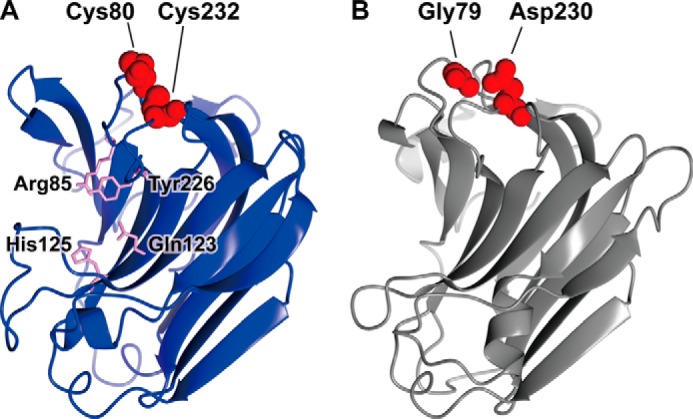
Homology modeling of NitAly and PyAly. The structures of NitAly (A, residues 34–242) and PyAly (B, residues 13–216) were predicted using the PHYRE2 program (63) and P. aeruginosa alginate lyase PA1167 (Protein Data Bank code 1vav) as the template (15) with 100% confidence, respectively. A, Cys-80 and Cys-232 are shown by red globules. The residues Arg-85, Gln-123, His-125, and Tyr-226, which correspond to the suggested catalytic residues in PA1167 (15, 21), are shown by pink sticks. B, Gly-79 and Asp-230 are shown by red globules.
