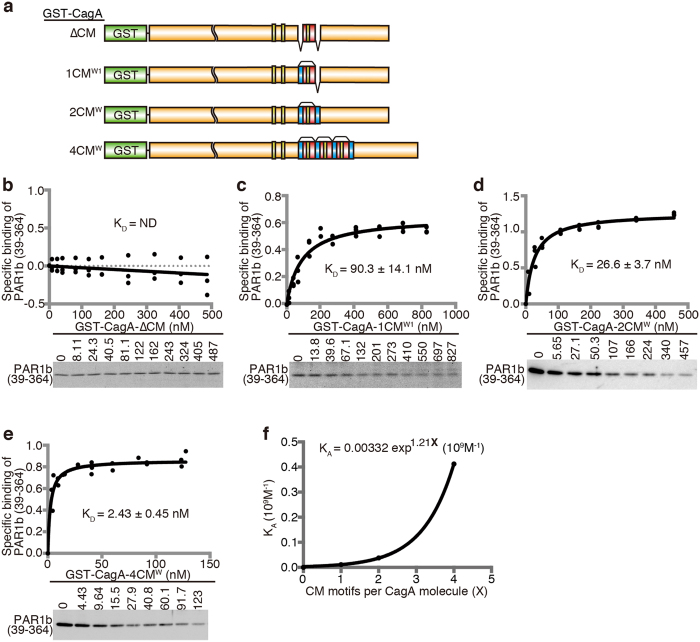
Figure 3. Tandem CM motifs generate strong binding affinity to PAR1b.
(a) Schematic diagram of GST-CagA constructs used. CagA-4CMW was derived from H. pylori NCTC11637 strain. (b–e) Saturation binding curves obtained by quantitative GST binding assay between PAR1b (39–364) and GST-CagA-ΔCM (b), GST-CagA-1CMW1 (c), GST-CagA-2CMW (d), or GST-CagA-4CMW (e). Values shown are dissociation constants (KD) ± SE, n = 3. Bands at the bottom of each graph shows a representative CBB stained gel or an immunoblot of unbound PAR1b (39–364). Full-length gels and blots are presented in Supplementary Fig. S2. (f) Graph and equation depicting the relationship between the number of CM motifs per molecule of CagA and their association constants (KA) with PAR1b (39–364).