Abstract
In aquatic environments, fungal communities remain little studied despite their taxonomic and functional diversity. To extend the ecological coverage of this group, we conducted an in-depth analysis of fungal sequences within our collection of 3.6 million V4 18S rRNA pyrosequences originating from 319 individual marine (including sea-ice) and freshwater samples from libraries generated within diverse projects studying Arctic and temperate biomes in the past decade. Among the ~1.7 million post-filtered reads of highest taxonomic and phylogenetic quality, 23,263 fungal sequences were identified. The overall mean proportion was 1.35%, but with large variability; for example, from 0.01 to 59% of total sequences for Arctic seawater samples. Almost all sample types were dominated by Chytridiomycota-like sequences, followed by moderate-to-minor contributions of Ascomycota, Cryptomycota and Basidiomycota. Species and/or strain richness was high, with many novel sequences and high niche separation. The affinity of the most common reads to phytoplankton parasites suggests that aquatic fungi deserve renewed attention for their role in algal succession and carbon cycling.
High-throughput sequencing methods have augmented our capacity to assess microbial eukaryotic diversity and related function in microbial ecology. In freshwater and marine environments, attention has often been placed on protist diversity including phytoplankton and heterotrophic flagellates1,2, but the fungal components are often set aside or given cursory analyses without considering finer taxonomic levels. This contrasts with microbial studies on soil environments, in which fungi are considered the primary agents of decomposition3. The lesser attention to aquatic fungi is perhaps due to their overall low abundances in marine clone libraries (~1% of total eukaryotes4) and a perception that they may therefore be of little ecological relevance. However, some studies have shown larger proportions of fungi5 and the taxonomic richness of fungi in marine environments is reported to be high6. One group especially, the Chytridiomycota, has been identified as an early divergent lineage implicated in a variety of ecological processes. The role of Chytridiomycota as parasites of freshwater algae is well known7,8 and taxa in this group are thought to mediate the transfer of organic matter from phytoplankton into zooplankton via saprophytic and parasitic activity described as the “mycoloop”9. Freshwater chytrids are also known as parasites of zooplankton10 and of larger animals such as amphibians11. However, Chytridiomycota are rarely reported from marine environments and there are few marine cultured isolates12. Based on studies to date, it has been suggested that Dikarya (Ascomycota and Basidiomycota) dominate marine fungi4,13 and hyphomycetes (mostly assigned to the Ascomycota) play a major role decomposing leaf litter and other terrigenous substrates in rivers and streams14.
Direct examination of fungal diversity using high-throughput sequencing has been increasingly applied13,14,15,16, but data are still lacking in many environments, especially pelagic and polar regions. To more broadly assess aquatic fungal diversity, we examined pyrosequencing libraries of the V4 region of 18S rRNA (DNA and RNA) generated within our research group’s 19 projects studying Arctic and temperate biomes in the past decade (Fig. 1). The database totaled more than 3.6 million sequences, originating from 319 individual marine, freshwater and sea-ice samples (0.2–50 μm size-fractions), from 103 locations sampled from 2003 to 2011 (Supplementary Table S1).
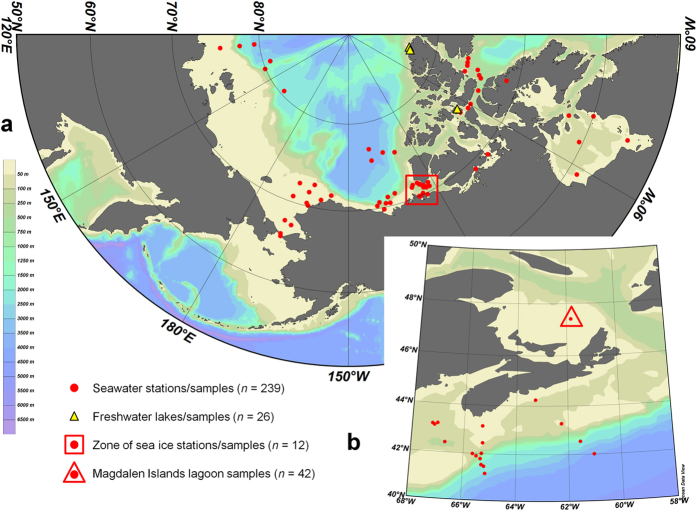
Figure 1. Locations of sampling sites included in pyrosequencing libraries.
(a) Arctic samples, ranging through Canadian, American and Russian territories. The box outlines the area of origin for the sea ice samples and the bathymetric scale at left applies to both maps. (b) Temperate samples from the Atlantic Ocean, including the Magdalen Islands lagoon samples (triangle). Not shown is the location of the Lake Tahoe samples (39°06’N 120°02’W). Maps created with Ocean Data View version 4 ( https://odv.awi.de/en/home/).
Results and Discussion
Our analysis showed the presence of fungi in all samples tested. The initial raw reads were reduced to ~1.7 million reads of highest taxonomic and phylogenetic quality using the filtering methods outlined below. Among these, 23,263 fungal sequences were identified, for an overall rate of incidence of 1.35%.
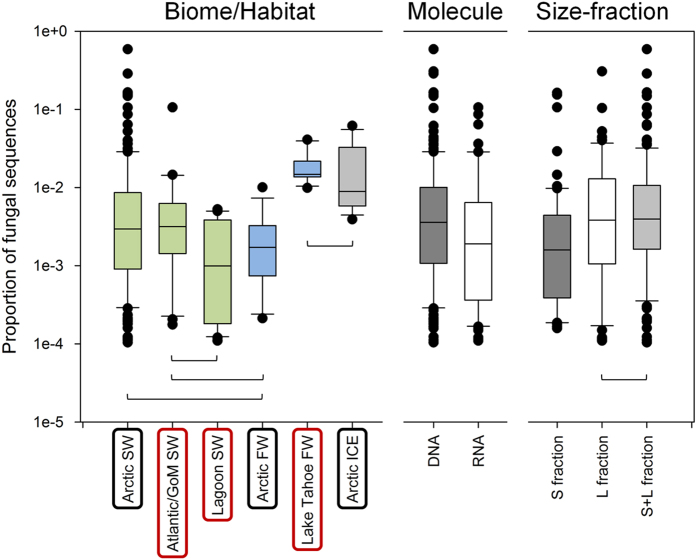
There were large variations among and within habitat types (Fig. 2), but the overall mean incidences were uniformly <5%. For example, there was large variability (0.01–59%) in Arctic seawater, which represented the greatest number of samples and overlapped with the ranges of other seawater habitats. There were, however, significantly higher proportions of fungi in sequences from temperate freshwater and Arctic sea-ice compared to the other habitats. There were also slightly higher fungal proportions in DNA relative to RNA (cDNA) libraries, possibly because resting stages, including spores, may have been more numerous among the fungal reads, leading to higher numbers in DNA signals. The higher proportion of fungal reads in the DNA derived libraries could also reflect the preservation of non-living DNA in cold waters17,18. Higher proportions of fungi occurred in the large size-fractions (>3 μm) that, in part, may reflect the physical association with detritus and nanoplankton. There were no significant correlations between the different fungal taxa and month, season, depth, temperature or salinity. However, there was a slight negative correlation between total fungi and temperature within the temperate samples, which were from sites spanning the largest temperature range, suggesting a preference for warmer water column conditions (Supplementary Fig. S1).
Figure 2. Incidences of fungal sequences.
Box-plots of proportions of fungal sequences in the different biomes/habitats (left panel), in DNA vs. RNA samples (middle panel) and in the different size fractions (right panel) – small (S), large (L) or when both were mixed (S + L). Boxes are significantly different from all other boxes in the same panels (Kruskal-Wallis [Bonferroni corrected] or Mann-Whitney, p < 0.05), unless linked by brackets. FW, freshwater; SW, seawater.
A total of 44 genera (plus one clone and one informal genus) of fungi were identified, contained within 9 (sub)phyla (including incertae sedis), a testament to the large richness of fungi that can be captured by the universal 18S primers. Although ITS regions are often used to examine species- and strain-level fungal diversity, 18S rRNA sequences have proven valuable in many fungal analyses13,19,20,21,22. In the present study, our moderately-sized (~440 bp) V4 amplicons resolved fungal sequences to at least the genus level, confirmed by manual BLASTn of taxonomic identifications. The full taxonomic breakdown of the samples is presented in Supplementary Table S2.
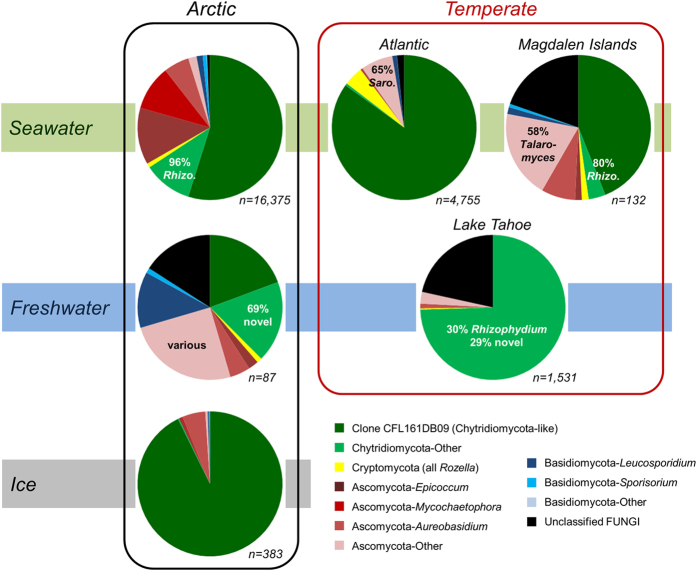
A striking overall feature of our fungal sequence dataset was that almost all sample types were dominated by Chytridiomycota-like sequences (38–93% for the groupings in Fig. 3). This was followed by moderate contributions of Ascomycota (4–32%) and relatively minor contributions of Cryptomycota (0–4%; all Rozella spp.) and Basidiomycota (0–14%). For the Basidiomycota, Leucosporidium was the most common genus (notably in Arctic freshwaters), whereas within the Ascomycota, Epicoccum, Mycochaetophora, Aureobasidium, Talaromyces and Sarocladium were recovered in varying proportions. The high dominance of chytrids in Arctic sea-ice (93%) agrees with a recent 18S V2-V3 Alaskan study showing 70–95% chytrids among fungal sequences from land-fast ice and underlying marine sediments22. In contrast, a recent meta-analysis by Tisthammer et al.13 focused on marine water and sediments found that Dikarya were dominant and chytrids were relatively rare. However, their study was based upon only 56 samples from 33 sites, identified less than half the number of fungal sequences as our study, and had a limited coverage of polar regions. They also targeted the small ~65 bp V9 variable region of the 18S rRNA gene and, consequently, greater than 50% of their 10,793 sequences remained unidentified. Our V4 analysis with a larger dataset over a broad range of aquatic environments, with emphasis on planktonic and sea-ice systems, implies that chytrids may be more abundant than previously suspected.
Figure 3. Taxonomic identities of fungal sequences in the various biomes/habitats.
The total numbers of sequences (n) in each category are indicated. Rhizo., Rhizophydium; Saro., Sarocladium.
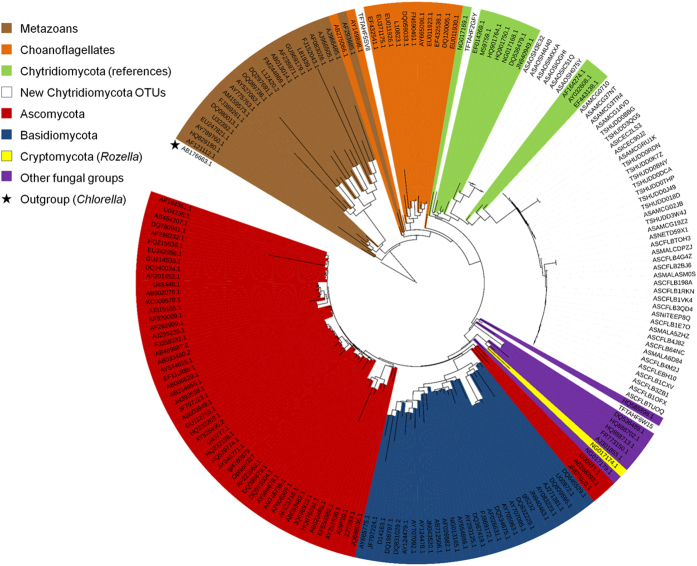
Within the Chytridiomycota, sequences with closest match to uncultured clone CFL161DB09 (HM561158.1) were by far dominant, except in Lake Tahoe where it was clone PA2009D12 (HQ191408) from meromictic Lake Pavin15, followed by Rhizophydium-like sequences. Clone CFL161DB09 originates from an Arctic seawater sample from the Amundsen Gulf and is representative of a small cluster of clones from the same study23. These Arctic clones, and the large set of additional sequences from this study, group together with known Chytridiomycota at the base of the fungal phylogenetic tree (Fig. 4). However, the closest cultured species, Clydaea vesicula (alternatively annotated as “Chytridiales sp.” JEL369 [EF443137] and PL70 [EF443138]) within the Lobulomycetales24, shares only ~91% identity, therefore these environmental sequences represent novel lineages of aquatic Chytridiomycota.
Figure 4. Phylogeny of the top 50 Chytridiomycota-like OTUs (98% identity) from all samples along with reference sequences.
This 18S maximum-likelihood tree of the V4 region only ( = 401–462 bp) was obtained with RAxML (with 100 bootstrap replicates) and rooted with a Chlorella 18S sequence (Viridiplantae; AB176663.1). Fungi color-coding matches Fig. 3. Reference sequences are identified by their NCBI accession numbers and novel sequences from this study are identified as follows: (A/T) + (F/S) + project + uniqueID; where A/T is Arctic vs. Temperate, F/S is Freshwater vs. Seawater, the project is identified with a 3-letter code (AMC = AMCE, AOS, CFL, HUD = Hudson, ICE = ICESCAPES, MAL = MALINA, NET = ArcticNet’11, NIT = ANITA, TAH = Lake Tahoe; Supplementary Table S1), and a 5-letter code identifies the representative sequence for the OTU (allowing for retrieval in Supplementary File S1).
To assess the overall diversity of the fungal sequences, they were clustered into Operational Taxonomic Units (OTUs) at 98% identity and a shared OTU analysis was conducted between all sample type combinations (latitude vs. source medium; Supplementary Fig. S2). We used the clustering technique as a tool allowing for these β-diversity comparisons between samples, however we add the caveat that the 98% level may not be strictly appropriate for all fungal groups; some may require higher identity, some less, to reflect accurate species-level clusters. Nonetheless, little overlap was observed between freshwater, seawater and sea-ice within the same biomes, indicating that species (or strain) richness was high and that environmental niche separation was strong. Only 168 OTUs (representing ~4% of the total set) were universally shared between the polar and temperate latitudes, and the majority of these were identified as CFL161DB09-like novel Chytridiomycota (Supplementary Fig. S3). Although shared, an examination of the distributions of the top 40 dominant OTUs (Supplementary Fig. S4) showed inverse sequence count patterns between the two biomes, indicating that each environment and aquatic medium was populated by its own distinct lineages or strains. Similar situations were recently encountered for land-based Chytridiomycota full-length 18S clones from high-elevation soils and snow19,20. Comparison of soil and aquatic Chytridiomycota was outside the scope of the present study, but such analysis in the future would yield interesting insights into the evolution of this group. High niche or biome compartmentalization was also observed in a meta-analysis of a small set of full-length 18S fungal sequences (~1,800) by Panzer et al.21 using coarse habitat definitions (e.g., all seawater samples grouped as one) and the Tisthammer et al.13 meta-analysis comparing pelagic and benthic communities.
The ecological roles of the novel lineages detected in the present study are unknown, however many chytrids, including numerous species within the genus Rhizophydium, have long been known as algal parasites7. Lepère et al.25 recently demonstrated direct in situ hybridization evidence, albeit relatively uncommon, of fungi associated (potentially parasitically) with marine photosynthetic picoeukaryotes using specific chytrid and general fungal probes. In our study, significant positive correlations (Supplementary Fig. S1) were found between the proportions of Chytridiomycota-like sequences and either total chlorophyll a concentrations or proportions of diatom sequences, albeit with an rs of 0.29. More detailed studies following species-specific blooms over time using quantitative PCR or fluorescence in situ hybridization techniques are clearly needed. This broad meta-analysis of aquatic fungal sequences highlights the importance of further examining the roles of Chytridiomycota in marine and freshwater environments through molecular methods, and the need to pursue cultivation of dominant lineages such as arctic clone CFL161DB09 for use in physiological and ecological studies.
Materials and Methods
Sample collection and pyrosequencing
Sample collection, nucleic acid extractions and pyrosequencing are described in associated publications (listed in Supplementary Table S1) and were similar for all data sets, including those listed as unpublished from our research group. Briefly, to ensure sufficient biomass collection without blocking filters, 1–10 L of seawater or freshwater, and 0.2–0.5 L of melted ice, were filtered sequentially into small (S = 0.2–3 μm) and large (L = >3 μm) size-fractions. DNA was extracted using a modified salt-based method and RNA was extracted using the RNeasy Mini Kit (Qiagen), including a DNase treatment and followed immediately by cDNA synthesis, as described in Charvet et al.26 (excluding the MDA step). The V4 region of the 18S rRNAs (RNA) and rRNA genes (DNA) were amplified, pooled and pyrosequenced as described in Comeau et al.27,28. For some samples and stations only one molecule (DNA or RNA) or one size-fraction (L or S) was sequenced, but for many both were done in parallel. In some cases, the size-fractions were either mixed 50:50 or in accordance with the corresponding chlorophyll a or cell count ratios measured in each fraction (Supplementary Table S1).
Read quality screening and selection for fungi
Quality filtering (and further analyses herein) was conducted using mothur ( www.mothur.org). The 319 samples contained a cumulative 3,651,419 raw reads which were quality filtered as described in Comeau et al.27,28, but with a more stringent requirement of a 400 bp minimum length in order to only retain reads of highest taxonomic utility and phylogenetic quality for this heavily identification-based study. This resulted in 1,717,227 filtered reads: 1,032,555 Arctic marine, 53,456 Arctic freshwater, 21,829 Arctic ice, 527,710 temperate marine and 81,677 temperate freshwater. Reads were then screened using a custom 18S database (see below) at the “phylum” (eukaryote “major group”) level to select fungal sequences. Cumulatively, 23,263 fungal sequences were retained among all samples.
OTU generation and taxonomic identification
The fungal sequences were merged, aligned and OTUs were formed at the 98% similarity level (furthest-neighbor method; Supplementary File S1) following the recommendation for using a maximum of 98% with 454 GS-FLX Titanium chemistry to avoid misidentifying V4 Eukarya tags29. This permitted construction of rank-abundance curves for each sample type (α-diversity; Supplementary Fig. S4) and enabled a shared OTU analysis to examine sample type overlap (β-diversity; Supplementary Figs S2 and S3). Final taxonomic identification of OTUs and sequences was conducted using an 18S database developed in-house, originally described in Comeau et al.27,28, but updated to be more inclusive of fungal diversity, including uncultured environmental clones30. This database was also further refined by an iterative process whereby major “unclassified” fungal sequences from this study were manually investigated by BLASTn in GenBank nr (blast.ncbi.nlm.nih.gov), new reference sequences were added (if found), and the classification process for all sequences was repeated until satisfactory identification was achieved.
Phylogenetic analysis
The top 50 Chytridiomycota-like OTUs were aligned with other fungal and metazoan reference sequences from GenBank, trimmed to only the V4 region (401–462 bp), using the implementation of ClustalW in BioEdit ( www.mbio.ncsu.edu/bioedit/bioedit.html). The eye-refined alignment was used to construct a maximum-likelihood tree in RAxML (sco.h-its.org/exelixis/web/software/raxml/) with 100 bootstrap replicates, rooted with a Chlorella sequence (Viridiplantae; AB176663.1). The tree was visualized and exported using the Interactive Tree of Life (iTOL; itol.embl.de).
Statistical analysis
Non-parametric (non-normal) Mann-Whitney and Kruskal-Wallis tests were used to verify differences between groups and Spearman’s r (on log-transformed data) was used for correlations. F- and t-tests (equal or unequal variance, as was the case) and linear regressions (log-transformed data) were used with data that fulfilled the conditions of normality. All statistical analyses were carried out with PAST (folk.uio.no/ohammer/past/).
Additional Information
How to cite this article: Comeau, André M. et al. Novel chytrid lineages dominate fungal sequences in diverse marine and freshwater habitats. Sci. Rep. 6, 30120; doi: 10.1038/srep30120 (2016).
Supplementary Material
Acknowledgments
We thank the various participants and crews of the CCGS Amundsen, CCGS Hudson, CCGS Louis St-Laurent and USCGC Healy for their logistical support during multiple research expeditions; and the following students working in the Lovejoy lab for generating additional pyrosequencing libraries mined in this work: Sophie Charvet, Cindy Dasilva, Marcos Lagunas, Ian Luddington, Emmanuelle Medrinal, Bérangère Péquin and Mary Thaler. Marianne Potvin contributed to pyrosequencing and technical supervision of students. The ICESCAPE DNA samples in the Chukchi Sea were collected and extracted by Eva Ortega-Retuerta. We thank Marcel Babin (the Canadian Excellence Research Chair in Remote Sensing of Canada’s New Arctic Frontier) for ICESCAPE access and Geoff Schladow for access to the sampling and laboratory facilities at the Tahoe Environmental Research Center, University of California Davis. We also thank our colleagues at the IBIS/Université Laval Plate-forme d’Analyses Génomiques for technical assistance. This work was funded by various grants from the Natural Sciences and Engineering Research Council (NSERC) of Canada and Fonds de recherche du Québec – Nature et technologies (FRQNT) supporting LB (CEF), WFV (CEN), and CL (Québec-Océan). The Magdalen Islands study was funded primarily by an NSERC Strategic Grant led by Gregor Fussman. We also thank Jill Watkins and the Department of Fisheries and Oceans, Canada for support for the Arctic Ocean Survey, which is a contribution to the Circumpolar Biodiversity Monitoring Plan. This is a contribution to the Canadian Healthy Oceans Network (CHONe, an NSERC Strategic Network) and ArcticNet (Canadian Network of Centres of Excellence).
Footnotes
Author Contributions A.M.C. conceived the study, carried out the primary analysis and wrote the paper. A.M.C., W.F.V., L.B. and C.L. analyzed the data and contributed to the final writing of the paper.
References
- Vaulot D., Eikrem W., Viprey M. & Moreau H. The diversity of small eukaryotic phytoplankton (<3 μm) in marine ecosystems. FEMS Microbiol. Rev. 32, 795–820 (2008). [DOI] [PubMed] [Google Scholar]
- Thaler M. & Lovejoy C. Biogeography of heterotrophic flagellate populations indicates the presence of generalist and specialist taxa in the Arctic Ocean. Appl. Environ. Microbiol. 81, 2137–2148 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardgett R. D. & van der Putten W. H. Belowground biodiversity and ecosystem functioning. Nature 15, 505–511 (2014). [DOI] [PubMed] [Google Scholar]
- Richards T. A., Jones M. D. M., Leonard G. & Bass D. Marine fungi: Their ecology and molecular diversity. Annu. Rev. Mar. Sci. 4, 495–522 (2012). [DOI] [PubMed] [Google Scholar]
- Orsi W., Song Y. C., Hallam S. & Edgcomb V. Effect of oxygen minimum zone formation on communities of marine protists. ISME J. 6, 1586–1601 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manohar C. S. & Raghukumar C. Fungal diversity from various marine habitats deduced through culture-independent studies. FEMS Microbiol. Lett. 341, 69–78 (2013). [DOI] [PubMed] [Google Scholar]
- Sparrow F. K. Aquatic Phycomycetes. (University of Michigan Press, 1943). [Google Scholar]
- Sime-Ngando T. Phytoplankton chytridiomycosis: fungal parasites of phytoplankton and their imprints on the food web dynamics. Front. Microbiol. 3, 361 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagami M., Miki T. & Takimoto G. Mycoloop: chytrids in aquatic food webs. Front. Microbiol. 5, 166 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redfield G. W. & Vincent W. F. Stages of infection and ecological effects of a fungal epidemic on the eggs of a limnetic copepod. Freshwat. Biol. 9, 503–510 (1979). [Google Scholar]
- Berger L. et al. History and recent progress on chytridiomycosis in amphibians. Fungal Ecol. 19, 89–99 (2016). [Google Scholar]
- Gleason F. H. et al. Zoosporic true fungi in marine ecosystems: A review. Mar. Fresh. Res. 62, 383–393 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tisthammer K. H., Cobian G. M. & Amend A. S. Global biogeography of marine fungi is shaped by the environment.Fungal Ecol. 19, 39–46 (2016). [Google Scholar]
- Bärlocher F. Aquatic hyphomycetes in a changing environment. Fungal Ecol. 19, 14–27 (2016). [Google Scholar]
- Monchy S. et al. Exploring and quantifying fungal diversity in freshwater lake ecosystems using rDNA cloning/sequencing and SSU tag pyrosequencing. Environ. Microbiol. 13, 1433–1453 (2011). [DOI] [PubMed] [Google Scholar]
- Monard C., Gantner S. & Stenlid J. Utilizing ITS1 and ITS2 to study environmental fungal diversity using pyrosequencing. FEMS Microbiol. Ecol. 84, 165–175 (2013). [DOI] [PubMed] [Google Scholar]
- Dupray E., Caprais M. P., Derrien A. & Fach P. Salmonella DNA persistence in natural seawaters using PCR analysis. J. Appl. Bacteriol. 82, 507–510 (1997). [DOI] [PubMed] [Google Scholar]
- Charvet S., Vincent W. F., Comeau A. M. & Lovejoy C. Pyrosequencing analysis of the protist communities in a High Arctic meromictic lake: DNA preservation and change. Front. Microbiol. 3, 422 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman K. R. et al. Evidence that chytrids dominate fungal communities in high-elevation soils. Proc. Natl. Acad. Sci. USA 106, 18315–18320 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naff C. S., Darcy J. L. & Schmidt S. K. Phylogeny and biogeography of an uncultured clade of snow chytrids. Environ. Microbiol. 15, 2672–2680 (2013). [DOI] [PubMed] [Google Scholar]
- Panzer K. et al. Identification of habitat-specific biomes of aquatic fungal communities using a comprehensive nearly full-length 18S rRNA dataset enriched with contextual data. PLoS ONE 10, e0134377 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassett B. T. & Gradinger R. Chytrids dominate arctic marine fungal communities. Environ. Microbiol. 18, 2001–2009 (2016). [DOI] [PubMed] [Google Scholar]
- Terrado R. et al. Protist community composition during spring in an Arctic flaw lead polynya. Polar Biol. 34, 1901–1914 (2011). [Google Scholar]
- Simmons D. R., James T. Y., Meyer A. F. & Longcore J. E. Lobulomycetales, a new order in the Chytridiomycota. Mycol. Res. 113, 450–460 (2009). [DOI] [PubMed] [Google Scholar]
- Lepère C., Ostrowski M., Hartmann M., Zubkov M. & Scanlan D. J. In situ associations between marine photosynthetic picoeukaryotes and potential parasites–a role for fungi? Environ. Microbiol. Rep. 8, 445– 451 (2015). [DOI] [PubMed] [Google Scholar]
- Charvet S., Vincent W. F. & Lovejoy C. Effects of light and prey availability on Arctic freshwater protist communities examined by high-throughput DNA and RNA sequencing. FEMS Microbiol. Ecol. 88, 550–564 (2014). [DOI] [PubMed] [Google Scholar]
- Comeau A. M., Li W. K. W., Tremblay J.-E., Carmack E. C. & Lovejoy C. Changes in Arctic Ocean microbial community structure following the 2007 record sea ice minimum. PLoS ONE 6, e27492 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comeau A. M. et al. Protists in Arctic drift and land-fast sea ice. J. Phycol. 49, 229–240 (2013). [DOI] [PubMed] [Google Scholar]
- Behnke A. et al. Depicting more accurate pictures of protistan community complexity using pyrosequencing of hypervariable SSU rRNA gene regions. Environ. Microbiol. 13, 340–349 (2011). [DOI] [PubMed] [Google Scholar]
- Lovejoy C., Comeau A. & Thaler M. Curated reference database of SSU rRNA for northern marine and freshwater communities of Archaea, Bacteria and microbial eukaryotes. v. 1.1. Nordicana D23, doi: 10.5885/45409XD-79A199B76BCC4110 (2016). [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.