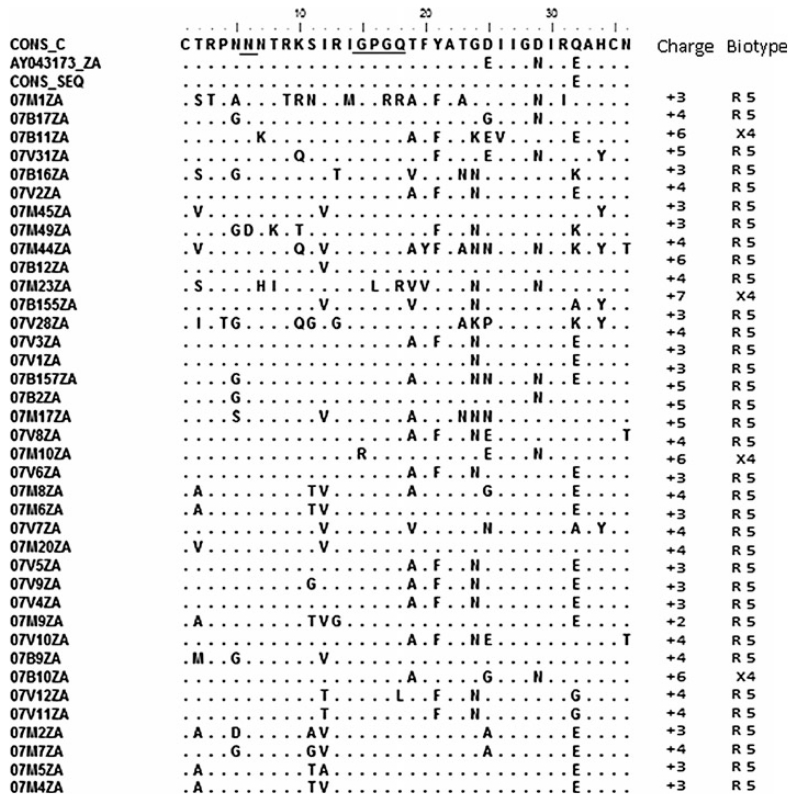
Fig. 4. Predicted V-3 loop amino acid sequences and biotypes of HIV from northeastern South Africa.
The multiple aligned sequences were compared to the consensus of the test sequences with those of the global subtype C consensus sequence and the South African vaccine strain AY043173_ZA. Underlined are the glycosylation sites and the GPGQ motif. Four viruses are X4 viruses, as predicted by webPSSM. The consensus of the test sequences showed close amino acid similarity to the global subtype C consensus sequence and the South African vaccine strain, except at position 32 for the consensus sequence and 25 and 29 for AY043173_ZA