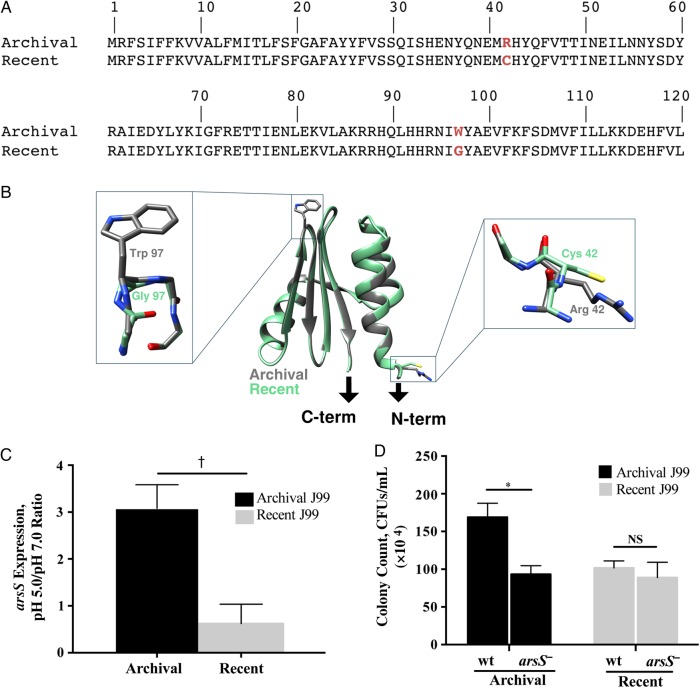
Figure 2.
The role of arsS in mediating acid survival in archival and recent Helicobacter pylori isolates. A, The arsS sequence from the archival J99 isolate was compared that from the recent J99 isolate, and 2 nonsynonymous point mutations were identified in the recent isolate. At amino acid position 42, cysteine was substituted for arginine, and at position 97, glycine was substituted for tryptophan (red). B, Ribbon diagram of a model of the ArsS sensor domain (residues 41–125). Arrows indicate connections to the putative transmembrane helices of ArsS. Side chains containing residues 42 and 97 are enlarged. C, Changes in H. pylori arsS expression in response to pH 5.0. RNA was isolated from H. pylori grown in broth adjusted to pH 5.0 or 7.0 using the RNeasy RNA purification kit (Qiagen). Reverse-transcriptase polymerase chain reaction (PCR) was performed using a High Capacity cDNA Reverse Transcription kit (Applied Biosystems), followed by reverse-transcriptase PCR using the SYBR Green Gene Expression Assay and a 7300 real-time PCR system (Applied Biosystems), with the following primers: arsS, 5′ AAGTGAAACGCTTCGCTCAAG 3′ and 5′ ATTCGTTAGCCAAATCCCCTATT 3′; 16S, 5′ TGCGAAGTGGAGCCAATCTT 3′ and 5′ GGAACGTATTCACCGCAACA 3′. Relative gene expression levels were calculated using the 2−ΔΔCT method. Levels of arsS expression at pH 5.0 and pH 7.0 were then normalized to levels of 16S messenger RNA expression, and data are represented as the ratio of pH 5.0 to pH 7.0; 3 independent experiments were conducted. †P = .003 (archival vs recent J99). D, Archival and recent wild-type (wt) J99 strains and their arsS isogenic mutants were cultured in Brucella broth that had been adjusted to a pH of 5.0 or 7.0 and then incubated for 60 minutes. Bacteria were then plated by serial dilution, and colonies were counted. *P = .02. Abbreviations: CFUs, colony-forming units; NS, not significant. This figure is available in black and white in print and in color online.