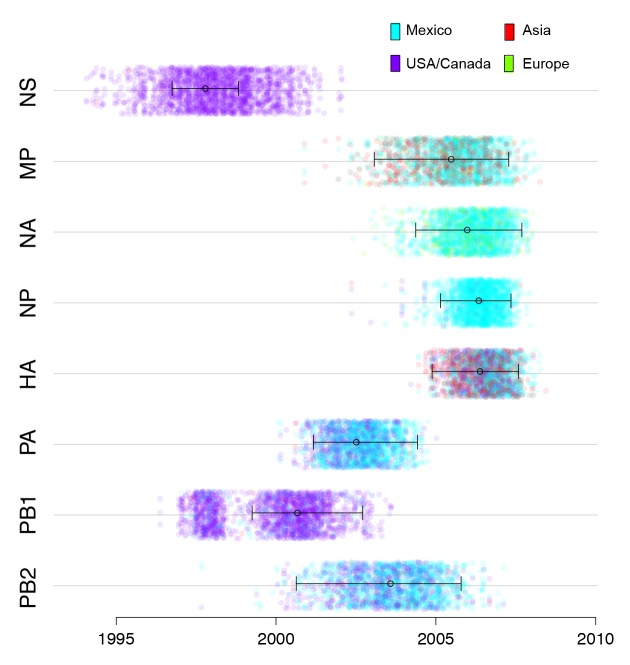
Figure 7. Timing and location of swine ancestors of pdmH1N1.
The color of each dot represents the inferred location of the node representing the common ancestor of pdmH1N1 viruses and the most closely related swine ivurses (indicated by open circles on the MCC trees in Figure 3), for a posterior distribution of ~2000 trees inferred for each segment. A high proportion of blue dots indicates a higher proportion of trees with Mexico as the inferred location state. The x-axis indicates the tMRCA of the same node, again for each tree. The 95% HPD is provided in brackets. Older tMRCAs are associated with longer phylogenetic branch lengths and gaps in sampling.
DOI: http://dx.doi.org/10.7554/eLife.16777.022