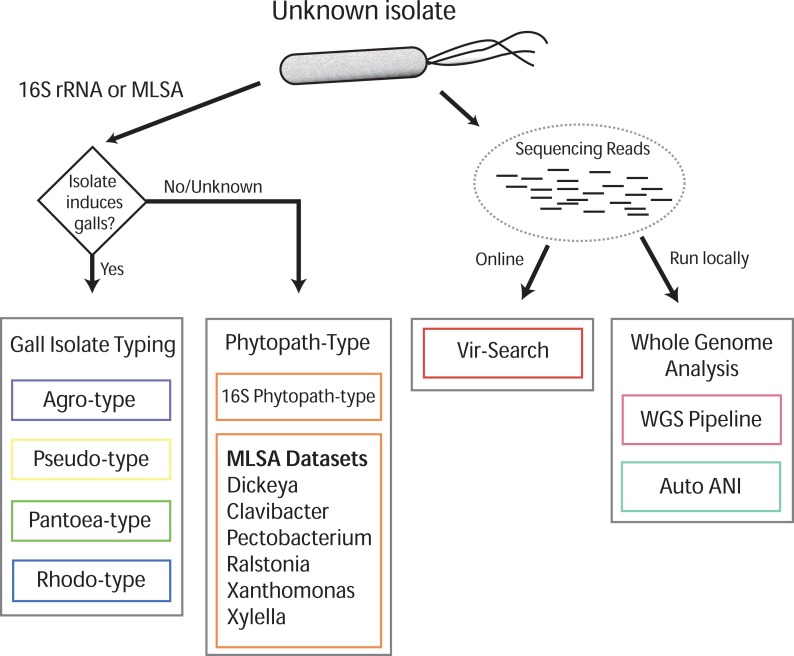
Figure 1. Overview of Gall-ID diagnostic tools.
DNA sequence information can be used to reveal the identity of the causative agent (unknown isolate) of disease. Tools associated with “Gall Isolate Typing” and “Phytopath-type” use 16S rDNA or pathogen-specific MLSA gene sequences to infer the identity of the isolate by comparing the sequences to manually curated sequence databases. Tools associated with “Whole Genome Analysis” and “Vir-Search” use Illumina short sequencing reads to characterize pathogenic isolates. The former tab provides downloadable tools to infer genetic relatedness based on SNPs (WGS Pipeline) or average nucleotide identity (Auto ANI). The “Vir-Search” tab provides an on-line tool to quickly map short reads against a database of sequences of virulence genes.