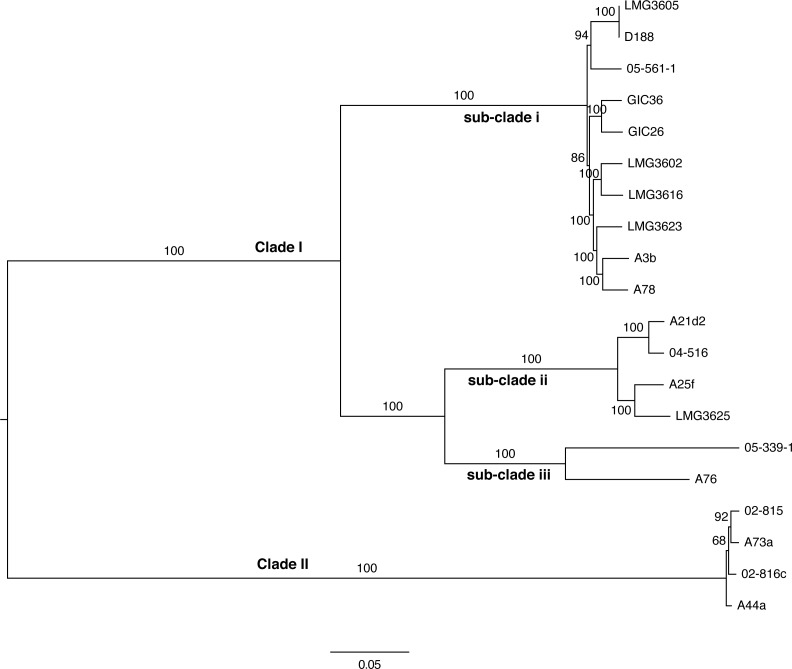
Figure 4. Maximum likelihood tree based on vertically inherited polymorphic sites core to 20 Rhodococcus isolates.
WGS Pipeline was used to automate the processing of paired end short reads from 20 previously sequenced Rhodococcus isolates, and generate a maximum likelihood unrooted tree. Sequencing reads were aligned, using R. fascians strain A44a as a reference. SNPs potentially acquired via recombination were removed. The tree is midpoint-rooted. Scale bar = 0.05 average substitutions per site; non-parametric bootstrap support as percentages are indicated for each node. Major clades and sub-clades are labeled in a manner consistent with previous labels.